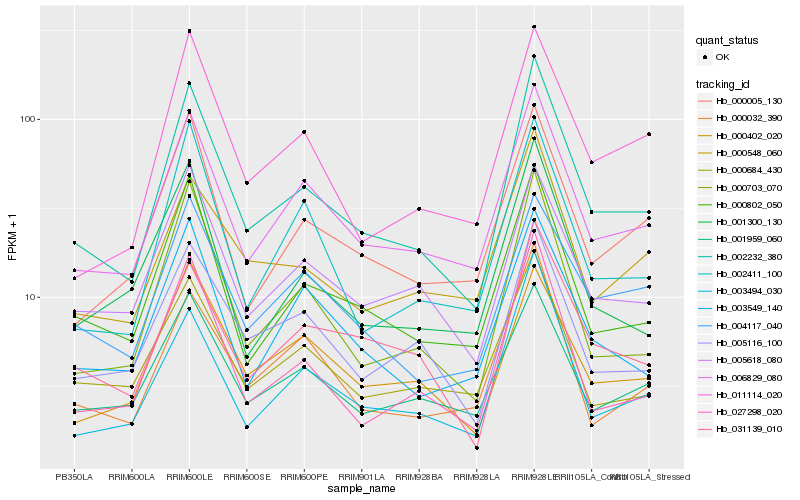
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_380 |
0.0 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_006829_080 |
0.0829746691 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 3 |
Hb_000802_050 |
0.0943312212 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 4 |
Hb_000703_070 |
0.0985388393 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 5 |
Hb_027298_020 |
0.1047047539 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 6 |
Hb_005618_080 |
0.1075534267 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003494_030 |
0.1090421943 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031139_010 |
0.1094925816 |
- |
- |
PREDICTED: uncharacterized protein LOC105632403 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005116_100 |
0.1119971211 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 10 |
Hb_000005_130 |
0.1134201687 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 11 |
Hb_011114_020 |
0.1148202436 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_001959_060 |
0.1157107392 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003549_140 |
0.1167897082 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001300_130 |
0.1173691522 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 15 |
Hb_002411_100 |
0.1176061797 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 16 |
Hb_000402_020 |
0.117718738 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000684_430 |
0.1197958396 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004117_040 |
0.1201154527 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 19 |
Hb_000548_060 |
0.1205814169 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 20 |
Hb_000032_390 |
0.1215541279 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |