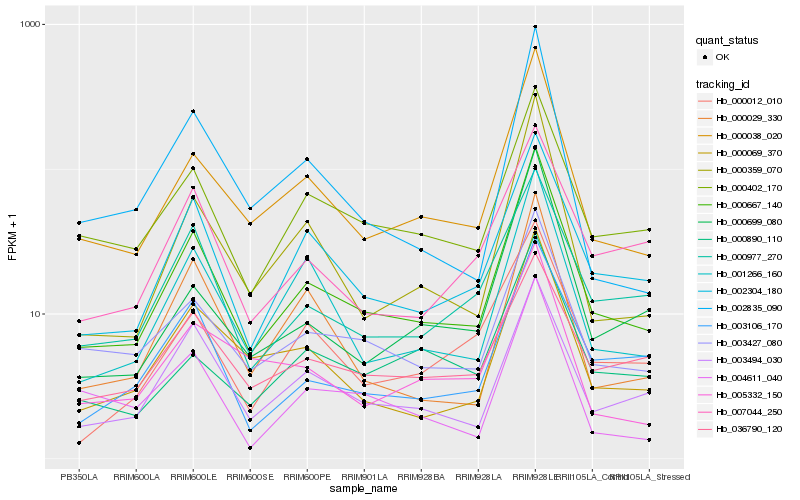
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000667_140 |
0.0 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000038_020 |
0.0837753482 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002304_180 |
0.0996025548 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003427_080 |
0.1083918546 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003494_030 |
0.1141401822 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001266_160 |
0.1145198672 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000359_070 |
0.116274453 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000402_170 |
0.117195462 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 9 |
Hb_000069_370 |
0.1178389787 |
- |
- |
- |
| 10 |
Hb_003106_170 |
0.1260181714 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 11 |
Hb_000977_270 |
0.1297000654 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000029_330 |
0.1361789893 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002835_090 |
0.1381462508 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 14 |
Hb_000012_010 |
0.1381965131 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 15 |
Hb_007044_250 |
0.1417233262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005332_150 |
0.1449995258 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 17 |
Hb_000890_110 |
0.1466937364 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 18 |
Hb_036790_120 |
0.1469539898 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000699_080 |
0.1473264957 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004611_040 |
0.1486068634 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |