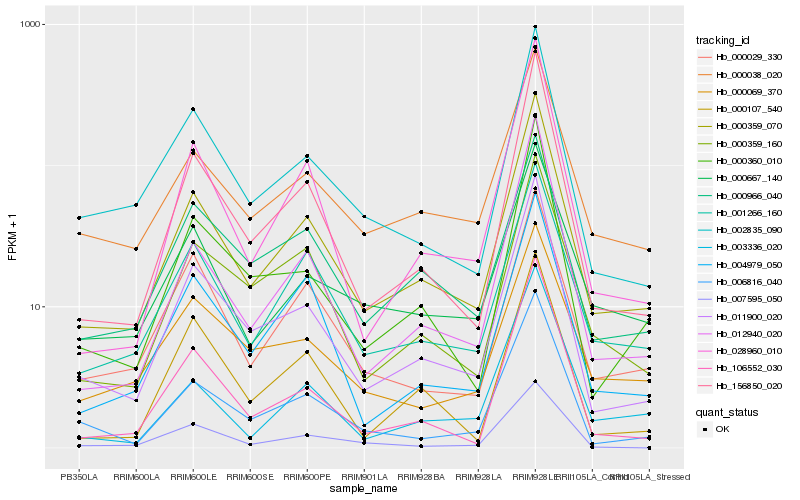
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_070 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_011900_020 |
0.0769323262 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 3 |
Hb_156850_020 |
0.0812588299 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 4 |
Hb_001266_160 |
0.0914818322 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_012940_020 |
0.097542563 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 6 |
Hb_000038_020 |
0.0986134809 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_028960_010 |
0.1012771462 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 8 |
Hb_000360_010 |
0.1082884606 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_002835_090 |
0.1102327897 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 10 |
Hb_000359_160 |
0.1119455147 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 11 |
Hb_004979_050 |
0.1128491911 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 12 |
Hb_006816_040 |
0.1128650589 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 13 |
Hb_000029_330 |
0.1132562833 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000667_140 |
0.116274453 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_106552_030 |
0.1217617765 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 16 |
Hb_003336_020 |
0.1292868081 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 17 |
Hb_000069_370 |
0.1296850272 |
- |
- |
- |
| 18 |
Hb_000966_040 |
0.1317974583 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 19 |
Hb_007595_050 |
0.1342824345 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 20 |
Hb_000107_540 |
0.1358281875 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |