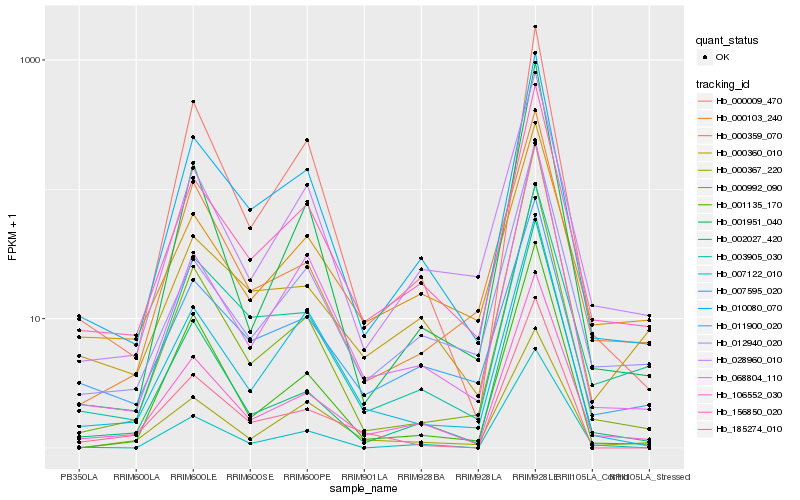
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106552_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 2 |
Hb_156850_020 |
0.0622294204 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 3 |
Hb_010080_070 |
0.0898572227 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000992_090 |
0.0939181653 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 5 |
Hb_012940_020 |
0.0968441823 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 6 |
Hb_068804_110 |
0.0990722856 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000360_010 |
0.1036538984 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_028960_010 |
0.1053945782 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 9 |
Hb_011900_020 |
0.1128093157 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002027_420 |
0.1155011501 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 11 |
Hb_000103_240 |
0.1165922877 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 1 [Jatropha curcas] |
| 12 |
Hb_003905_030 |
0.1184102127 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_000009_470 |
0.1197764343 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000359_070 |
0.1217617765 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_185274_010 |
0.1227190266 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 16 |
Hb_001135_170 |
0.1227290783 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 17 |
Hb_007595_020 |
0.1227570883 |
- |
- |
maturase K [Hevea brasiliensis] |
| 18 |
Hb_000367_220 |
0.1229549618 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 19 |
Hb_001951_040 |
0.1246661687 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_007122_010 |
0.1253746111 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |