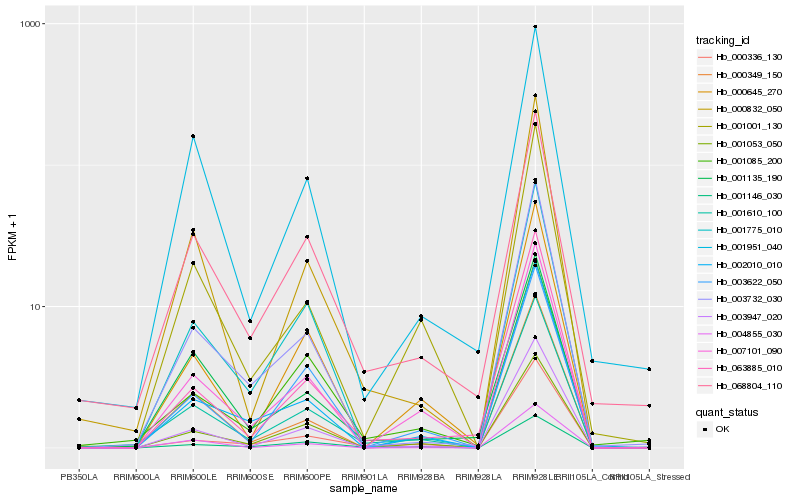
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001610_100 |
0.0 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000832_050 |
0.0852211415 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Marila laxiflora] |
| 3 |
Hb_007101_090 |
0.0864306218 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 4 |
Hb_003947_020 |
0.0938169504 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 5 |
Hb_068804_110 |
0.0944636333 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_003732_030 |
0.0974108008 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 7 |
Hb_001001_130 |
0.101197301 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 8 |
Hb_002010_010 |
0.1042901237 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 9 |
Hb_000336_130 |
0.1043793079 |
- |
- |
Triacylglycerol lipase 2 precursor, putative [Ricinus communis] |
| 10 |
Hb_001053_050 |
0.1060962655 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 11 |
Hb_004855_030 |
0.1062492564 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_000645_270 |
0.1087366892 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 13 |
Hb_001951_040 |
0.1088583525 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001775_010 |
0.110051815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001146_030 |
0.1110004243 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_001135_190 |
0.1128144321 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 17 |
Hb_001085_200 |
0.1129665124 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 18 |
Hb_000349_150 |
0.1138224159 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 19 |
Hb_063885_010 |
0.1140197848 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_003622_050 |
0.1159087311 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |