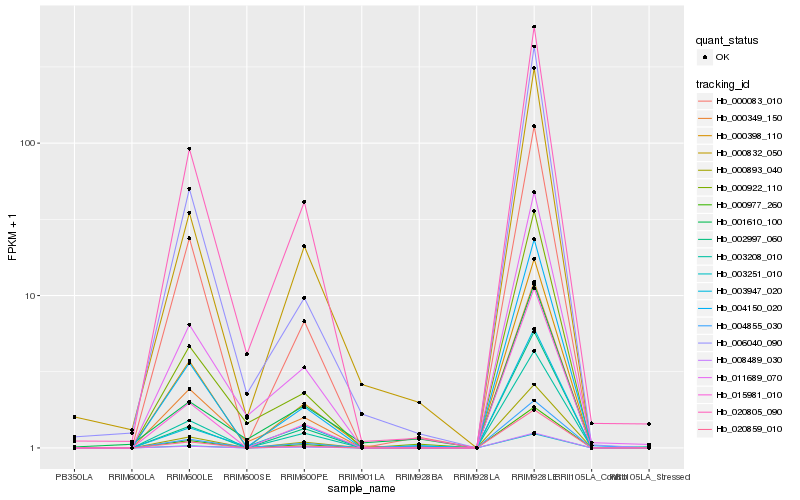
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000832_050 |
0.0 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Marila laxiflora] |
| 2 |
Hb_004855_030 |
0.0671157609 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_000349_150 |
0.0754715396 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 4 |
Hb_000893_040 |
0.0758017616 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 5 |
Hb_008489_030 |
0.0767170396 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_000977_260 |
0.0809684138 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 7 |
Hb_003251_010 |
0.0810300036 |
- |
- |
hypothetical protein MIMGU_mgv1a025126mg [Erythranthe guttata] |
| 8 |
Hb_004150_020 |
0.0813835668 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 9 |
Hb_020805_090 |
0.0820641254 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_002997_060 |
0.0830953361 |
- |
- |
hypothetical protein POPTR_0011s10730g [Populus trichocarpa] |
| 11 |
Hb_000398_110 |
0.0845846419 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 12 |
Hb_015981_010 |
0.0847045946 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_001610_100 |
0.0852211415 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_003208_010 |
0.0879154202 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 15 |
Hb_020859_010 |
0.0899856546 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X6 [Populus euphratica] |
| 16 |
Hb_003947_020 |
0.0903168942 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 17 |
Hb_011689_070 |
0.0930457615 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000922_110 |
0.0932826052 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 19 |
Hb_006040_090 |
0.0958686688 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Hevea brasiliensis] |
| 20 |
Hb_000083_010 |
0.0981895423 |
- |
- |
- |