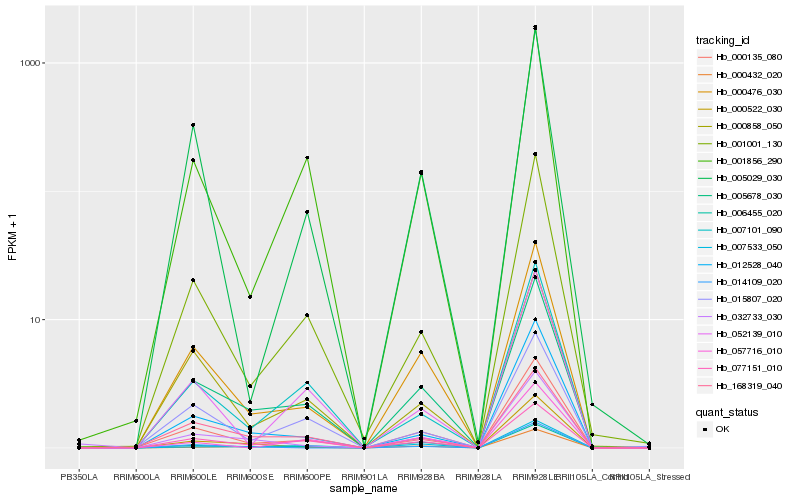
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000135_080 |
0.0 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_000476_030 |
0.0550878346 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 3 |
Hb_005678_030 |
0.0723697326 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 4 |
Hb_001001_130 |
0.0943853907 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 5 |
Hb_001856_290 |
0.103640502 |
- |
- |
hypothetical protein JCGZ_16255 [Jatropha curcas] |
| 6 |
Hb_012528_040 |
0.1040353873 |
- |
- |
- |
| 7 |
Hb_000858_050 |
0.1090643152 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_005029_030 |
0.1110315953 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |
| 9 |
Hb_000522_030 |
0.1133325464 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 10 |
Hb_077151_010 |
0.1161048027 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 11 |
Hb_007101_090 |
0.1214324752 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 12 |
Hb_052139_010 |
0.1233688166 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 13 |
Hb_015807_020 |
0.125524854 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 14 |
Hb_168319_040 |
0.1263947242 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 15 |
Hb_006455_020 |
0.1275184369 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_007533_050 |
0.138282577 |
- |
- |
- |
| 17 |
Hb_014109_020 |
0.1387467962 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 18 |
Hb_032733_030 |
0.1410726944 |
- |
- |
hypothetical protein CISIN_1g048657mg, partial [Citrus sinensis] |
| 19 |
Hb_057716_010 |
0.143396634 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_000432_020 |
0.1487655823 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |