| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
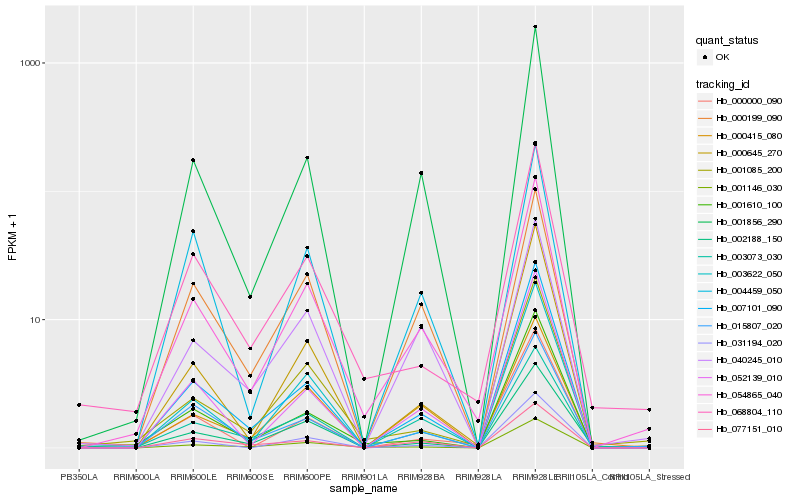
Hb_054865_040 |
0.0 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 2 |
Hb_001856_290 |
0.0949859659 |
- |
- |
hypothetical protein JCGZ_16255 [Jatropha curcas] |
| 3 |
Hb_015807_020 |
0.1026883203 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 4 |
Hb_001146_030 |
0.1092315515 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_007101_090 |
0.109919376 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 6 |
Hb_001085_200 |
0.110606389 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 7 |
Hb_052139_010 |
0.1153260422 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 8 |
Hb_000415_080 |
0.1158275844 |
- |
- |
- |
| 9 |
Hb_040245_010 |
0.1158415867 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |
| 10 |
Hb_068804_110 |
0.1171721962 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_004459_050 |
0.1197146635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003622_050 |
0.1207265067 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 13 |
Hb_000645_270 |
0.1212624163 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 14 |
Hb_003073_030 |
0.1213568972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_031194_020 |
0.1229171262 |
- |
- |
BnaCnng13030D [Brassica napus] |
| 16 |
Hb_077151_010 |
0.1230200256 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 17 |
Hb_001610_100 |
0.1235686661 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000000_090 |
0.1241071377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002188_150 |
0.1243736329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000199_090 |
0.1250787175 |
- |
- |
Tetrapyrrole-binding family protein [Populus trichocarpa] |