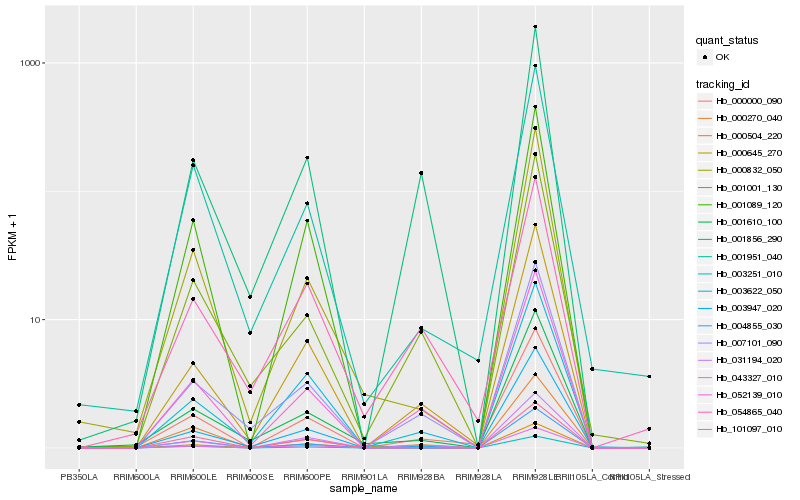
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_031194_020 |
0.0842962867 |
- |
- |
BnaCnng13030D [Brassica napus] |
| 3 |
Hb_000645_270 |
0.0857175596 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 4 |
Hb_052139_010 |
0.087462313 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 5 |
Hb_000270_040 |
0.10305847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007101_090 |
0.111206255 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 7 |
Hb_003622_050 |
0.1118792944 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 8 |
Hb_003947_020 |
0.1134807738 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 9 |
Hb_004855_030 |
0.1138250129 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 10 |
Hb_101097_010 |
0.1141143236 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 11 |
Hb_001951_040 |
0.1147516038 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000832_050 |
0.1190037111 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Marila laxiflora] |
| 13 |
Hb_001610_100 |
0.1195255585 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001001_130 |
0.1223272058 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 15 |
Hb_001856_290 |
0.1224696645 |
- |
- |
hypothetical protein JCGZ_16255 [Jatropha curcas] |
| 16 |
Hb_054865_040 |
0.1241071377 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 17 |
Hb_043327_010 |
0.1245858086 |
- |
- |
PREDICTED: uncharacterized protein LOC105785093, partial [Gossypium raimondii] |
| 18 |
Hb_003251_010 |
0.1250870575 |
- |
- |
hypothetical protein MIMGU_mgv1a025126mg [Erythranthe guttata] |
| 19 |
Hb_001089_120 |
0.1251259608 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 20 |
Hb_000504_220 |
0.1260210628 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |