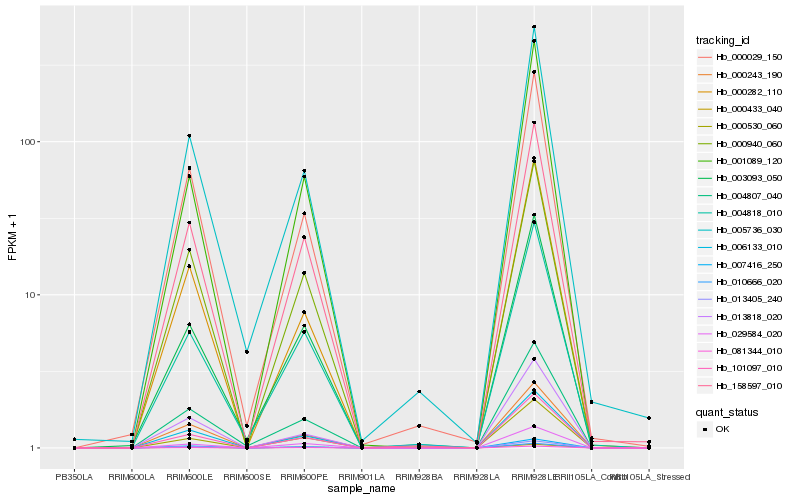
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101097_010 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_004807_040 |
0.0599284096 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 3 |
Hb_013405_240 |
0.0648303732 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 4 |
Hb_007416_250 |
0.065767036 |
- |
- |
- |
| 5 |
Hb_000530_060 |
0.065932851 |
- |
- |
- |
| 6 |
Hb_000029_150 |
0.0675396642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_158597_010 |
0.0687290739 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_005736_030 |
0.0695765045 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 9 |
Hb_001089_120 |
0.0695821285 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 10 |
Hb_029584_020 |
0.0703077219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000282_110 |
0.0749824655 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 12 |
Hb_003093_050 |
0.0753357018 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 13 |
Hb_081344_010 |
0.0756283271 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 14 |
Hb_000243_190 |
0.076511414 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 15 |
Hb_000433_040 |
0.076743463 |
- |
- |
- |
| 16 |
Hb_004818_010 |
0.0769272961 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 17 |
Hb_000940_060 |
0.0774665133 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 18 |
Hb_006133_010 |
0.078300502 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_013818_020 |
0.0801379381 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 20 |
Hb_010666_020 |
0.0826082038 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |