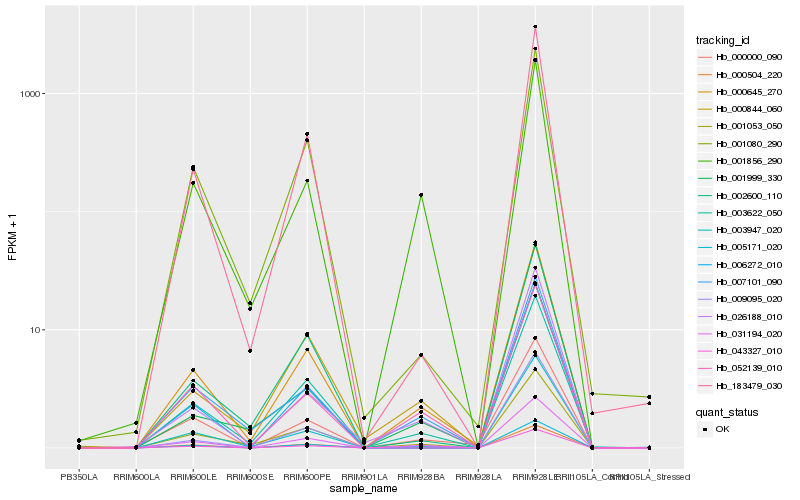
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031194_020 |
0.0 |
- |
- |
BnaCnng13030D [Brassica napus] |
| 2 |
Hb_000645_270 |
0.0460459502 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 3 |
Hb_003622_050 |
0.0653711603 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 4 |
Hb_052139_010 |
0.0704968883 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 5 |
Hb_000000_090 |
0.0842962867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007101_090 |
0.0936071029 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 7 |
Hb_000844_060 |
0.095858725 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 8 |
Hb_001856_290 |
0.0980415384 |
- |
- |
hypothetical protein JCGZ_16255 [Jatropha curcas] |
| 9 |
Hb_003947_020 |
0.1012493256 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 10 |
Hb_183479_030 |
0.1029421562 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001053_050 |
0.104808431 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 12 |
Hb_009095_020 |
0.1086921396 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 13 |
Hb_026188_010 |
0.1101541299 |
- |
- |
PREDICTED: tropinone reductase homolog [Jatropha curcas] |
| 14 |
Hb_001999_330 |
0.1117159857 |
- |
- |
hypothetical protein PRUPE_ppa022262mg, partial [Prunus persica] |
| 15 |
Hb_043327_010 |
0.1120913126 |
- |
- |
PREDICTED: uncharacterized protein LOC105785093, partial [Gossypium raimondii] |
| 16 |
Hb_005171_020 |
0.1130249359 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 17 |
Hb_000504_220 |
0.1154082238 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 18 |
Hb_006272_010 |
0.1166944786 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 19 |
Hb_002600_110 |
0.1180595026 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 20 |
Hb_001080_290 |
0.1184288824 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |