| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
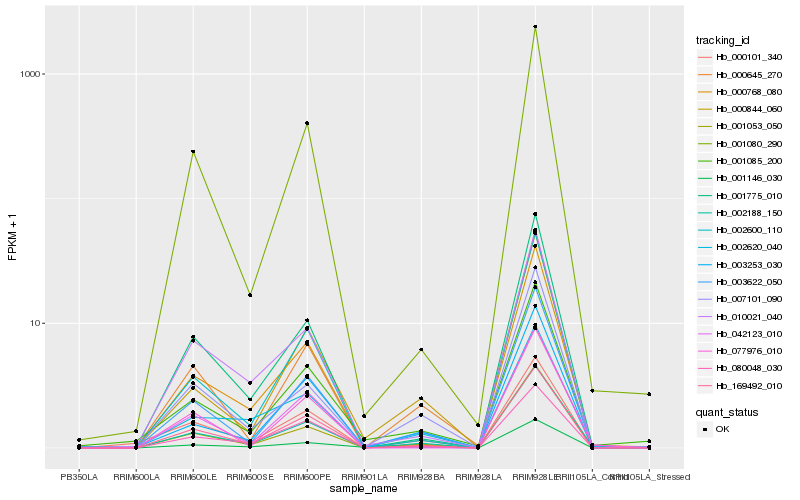
Hb_002188_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003622_050 |
0.0797393745 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 3 |
Hb_169492_010 |
0.0857251906 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 4 |
Hb_001053_050 |
0.0958976456 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 5 |
Hb_001775_010 |
0.1038652904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_077976_010 |
0.1067684815 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 7 |
Hb_001146_030 |
0.1082540037 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_000768_080 |
0.1083119394 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 9 |
Hb_001080_290 |
0.1085229003 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 10 |
Hb_002620_040 |
0.1088869574 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 11 |
Hb_001085_200 |
0.110026144 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 12 |
Hb_042123_010 |
0.1111285216 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 13 |
Hb_010021_040 |
0.1117800083 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 14 |
Hb_003253_030 |
0.1124834031 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_000844_060 |
0.1156348391 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 16 |
Hb_000645_270 |
0.1163559914 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 17 |
Hb_002600_110 |
0.1164272907 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 18 |
Hb_007101_090 |
0.1165314069 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 19 |
Hb_080048_030 |
0.1190645801 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 20 |
Hb_000101_340 |
0.1201201662 |
- |
- |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Ricinus communis] |