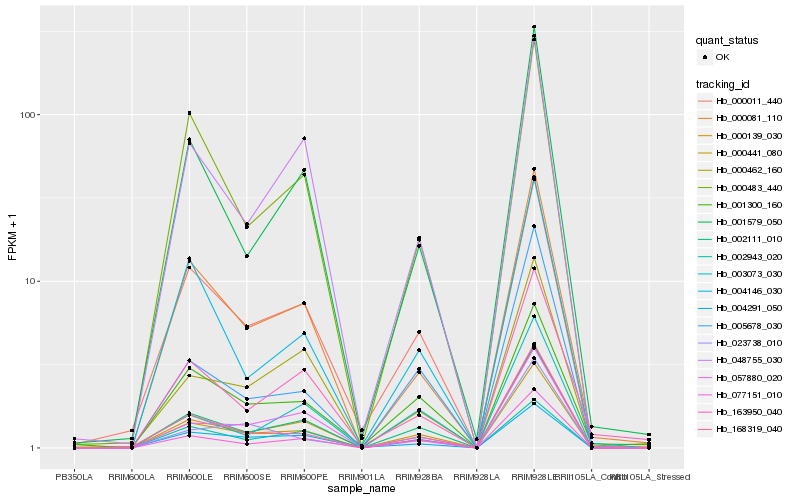
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_080 |
0.0 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 2 |
Hb_002111_010 |
0.0425816304 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 3 |
Hb_077151_010 |
0.0772780454 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 4 |
Hb_168319_040 |
0.091582701 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 5 |
Hb_000139_030 |
0.0917977935 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 6 |
Hb_000011_440 |
0.0942128993 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 7 |
Hb_000081_110 |
0.1011913604 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 8 |
Hb_001579_050 |
0.1100380835 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_005678_030 |
0.1122602248 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 10 |
Hb_048755_030 |
0.1131724577 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_057880_020 |
0.1210215528 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 12 |
Hb_000483_440 |
0.1235160297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000462_160 |
0.1239033986 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_003073_030 |
0.1287377654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004146_030 |
0.1289565951 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_023738_010 |
0.1291510648 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 17 |
Hb_001300_160 |
0.1298203504 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 18 |
Hb_002943_020 |
0.1351369952 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 19 |
Hb_163950_040 |
0.136025214 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004291_050 |
0.1361393525 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |