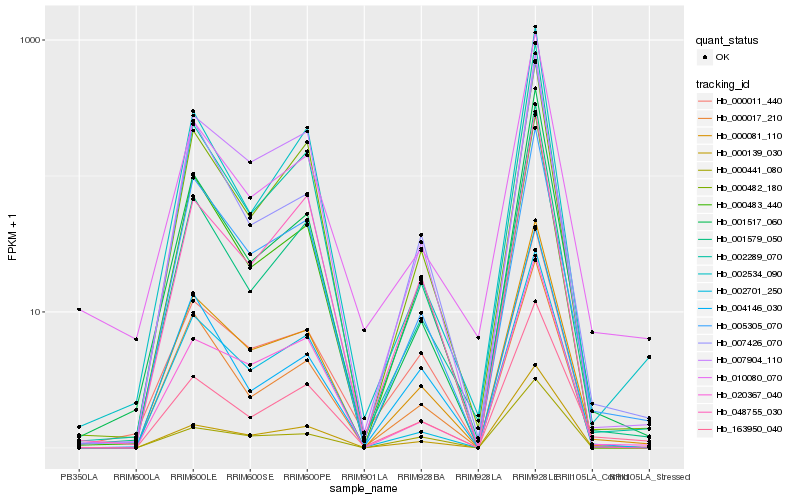
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000081_110 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_000483_440 |
0.0762542943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001517_060 |
0.0781081288 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 4 |
Hb_007426_070 |
0.0822373008 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 5 |
Hb_000139_030 |
0.0832477316 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 6 |
Hb_001579_050 |
0.083888304 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000017_210 |
0.0843872429 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 8 |
Hb_020367_040 |
0.0882334636 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 9 |
Hb_005305_070 |
0.0887018073 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 10 |
Hb_002701_250 |
0.0914099332 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 11 |
Hb_002289_070 |
0.0953831564 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000482_180 |
0.0971226403 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 13 |
Hb_163950_040 |
0.0980250405 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_048755_030 |
0.100927143 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_000441_080 |
0.1011913604 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 16 |
Hb_004146_030 |
0.1032995305 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_002534_090 |
0.1051513324 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 18 |
Hb_007904_110 |
0.1053885062 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000011_440 |
0.1058237267 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 20 |
Hb_010080_070 |
0.1060275025 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |