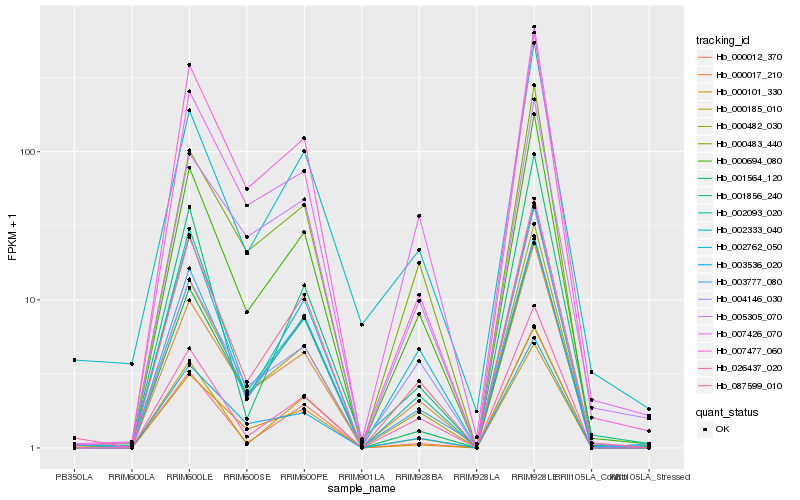
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000694_080 |
0.0 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 2 |
Hb_000017_210 |
0.0429685653 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 3 |
Hb_000482_030 |
0.048428688 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 4 |
Hb_087599_010 |
0.0526739159 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 5 |
Hb_007426_070 |
0.066584677 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 6 |
Hb_000483_440 |
0.0680858125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000012_370 |
0.0818667001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003536_020 |
0.0876153205 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 9 |
Hb_002762_050 |
0.0882752816 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 10 |
Hb_000101_330 |
0.0917267786 |
- |
- |
- |
| 11 |
Hb_003777_080 |
0.0924163261 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 12 |
Hb_004146_030 |
0.0926692814 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001564_120 |
0.0935582361 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 14 |
Hb_000185_010 |
0.0936364934 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 15 |
Hb_005305_070 |
0.0948268514 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 16 |
Hb_007477_060 |
0.0961165235 |
- |
- |
unknown [Medicago truncatula] |
| 17 |
Hb_002333_040 |
0.0973684581 |
- |
- |
hypothetical protein CISIN_1g015895mg [Citrus sinensis] |
| 18 |
Hb_026437_020 |
0.0976325536 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_001856_240 |
0.0977483628 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 20 |
Hb_002093_020 |
0.0978289192 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |