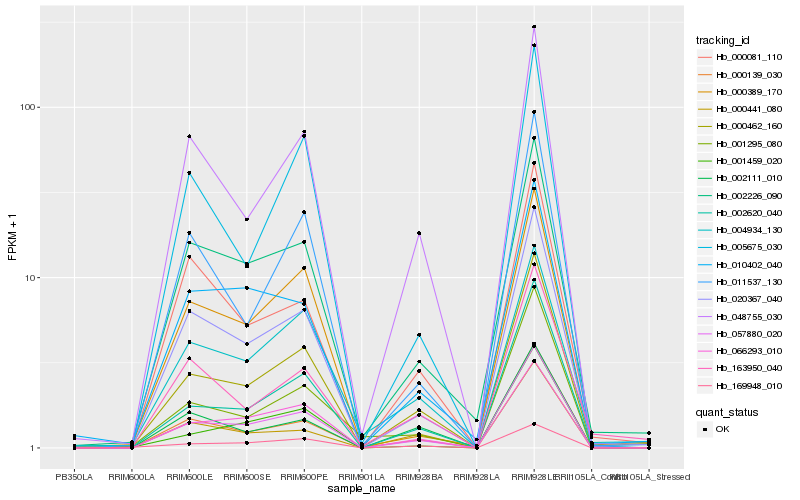
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000462_160 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_057880_020 |
0.0661598164 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 3 |
Hb_002620_040 |
0.0707282489 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 4 |
Hb_020367_040 |
0.0905668323 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 5 |
Hb_000139_030 |
0.091002533 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 6 |
Hb_048755_030 |
0.0953790119 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_002226_090 |
0.1091144459 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 8 |
Hb_004934_130 |
0.1158303233 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 9 |
Hb_163950_040 |
0.118181026 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011537_130 |
0.1210575319 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002111_010 |
0.1212564663 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 12 |
Hb_066293_010 |
0.1237274535 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_000441_080 |
0.1239033986 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_169948_010 |
0.1245654102 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 15 |
Hb_001295_080 |
0.1246087126 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 16 |
Hb_005675_030 |
0.1263796522 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 17 |
Hb_000389_170 |
0.1279378027 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 18 |
Hb_010402_040 |
0.1297566154 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 19 |
Hb_000081_110 |
0.131227183 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 20 |
Hb_001459_020 |
0.1318983463 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |