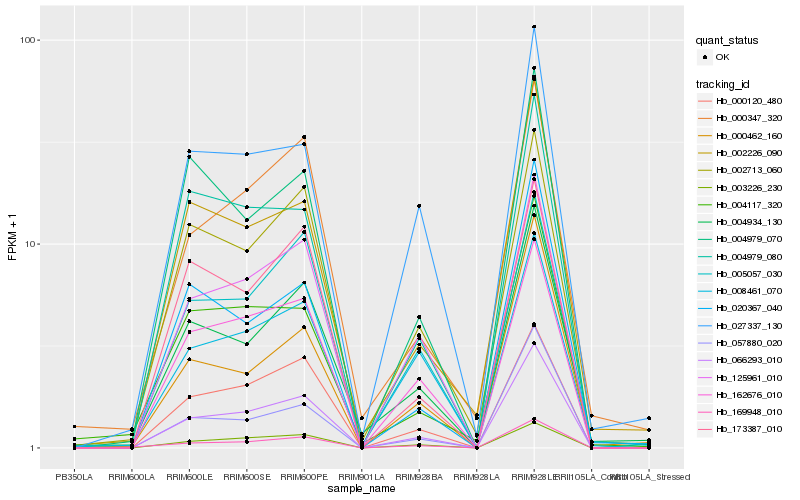
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066293_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 2 |
Hb_125961_010 |
0.0735746852 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_003226_230 |
0.0877413985 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 4 |
Hb_169948_010 |
0.0898545355 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 5 |
Hb_000120_480 |
0.0926883348 |
- |
- |
- |
| 6 |
Hb_162676_010 |
0.0973157569 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_057880_020 |
0.1014270361 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 8 |
Hb_004934_130 |
0.1068318831 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 9 |
Hb_002713_060 |
0.1086738424 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 10 |
Hb_004979_080 |
0.1107186993 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_027337_130 |
0.1121546608 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 12 |
Hb_005057_030 |
0.1140190377 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_000347_320 |
0.1156034989 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 14 |
Hb_002226_090 |
0.1166086818 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 15 |
Hb_173387_010 |
0.1192402522 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 16 |
Hb_004979_070 |
0.1216949697 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 17 |
Hb_008461_070 |
0.123127401 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000462_160 |
0.1237274535 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_020367_040 |
0.1240802404 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 20 |
Hb_004117_320 |
0.1250410224 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |