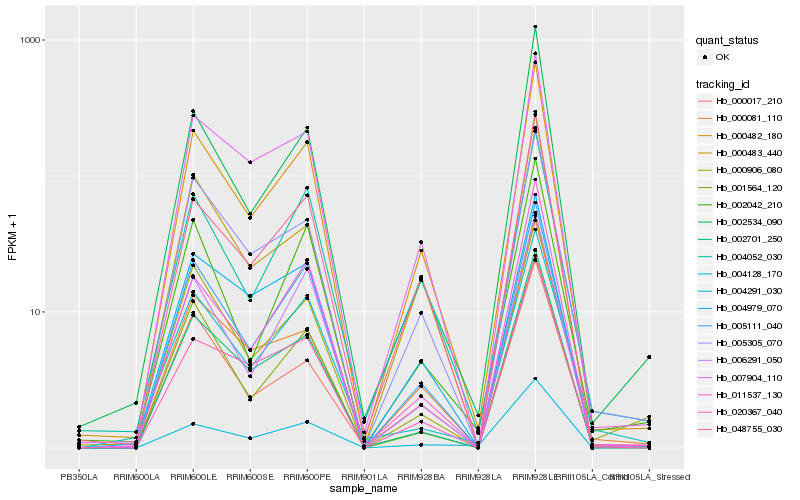
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000482_180 |
0.0 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 2 |
Hb_005111_040 |
0.0551571577 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 3 |
Hb_048755_030 |
0.0648803827 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_002701_250 |
0.0775492254 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_007904_110 |
0.0811709355 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_006291_050 |
0.0851874646 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_005305_070 |
0.0854270852 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 8 |
Hb_000483_440 |
0.089520379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004979_070 |
0.0905624649 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 10 |
Hb_002534_090 |
0.0906366157 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 11 |
Hb_004052_030 |
0.0909251148 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 12 |
Hb_001564_120 |
0.0926697053 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 13 |
Hb_002042_210 |
0.0934340029 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004291_030 |
0.0957177873 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 15 |
Hb_000017_210 |
0.0958497964 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 16 |
Hb_020367_040 |
0.0961742611 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 17 |
Hb_011537_130 |
0.0969704757 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000081_110 |
0.0971226403 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_004128_170 |
0.0978482972 |
- |
- |
hypothetical protein POPTR_0016s08350g [Populus trichocarpa] |
| 20 |
Hb_000906_080 |
0.0978590705 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |