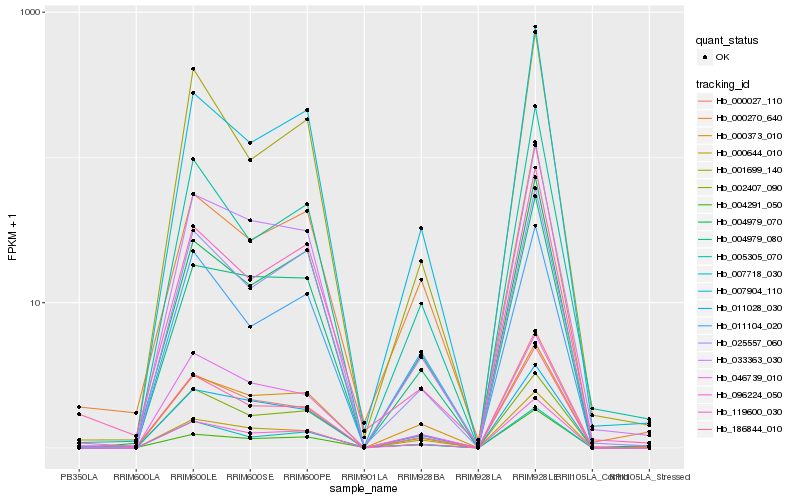
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096224_050 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 2 |
Hb_007904_110 |
0.0588555516 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_004979_070 |
0.059252212 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 4 |
Hb_000644_010 |
0.0622977358 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004979_080 |
0.0644435594 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000027_110 |
0.0684354474 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 7 |
Hb_033363_030 |
0.0687699783 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 8 |
Hb_025557_060 |
0.0757819498 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_000373_010 |
0.0792924215 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_002407_090 |
0.0818245168 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 11 |
Hb_007718_030 |
0.0892093556 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_011028_030 |
0.0897564166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005305_070 |
0.0908695839 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 14 |
Hb_186844_010 |
0.0920336774 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 15 |
Hb_001699_140 |
0.0957404116 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 16 |
Hb_046739_010 |
0.0970137477 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000270_640 |
0.09833366 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 18 |
Hb_119600_030 |
0.0991632567 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 19 |
Hb_011104_020 |
0.0995533596 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 20 |
Hb_004291_050 |
0.1003333342 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |