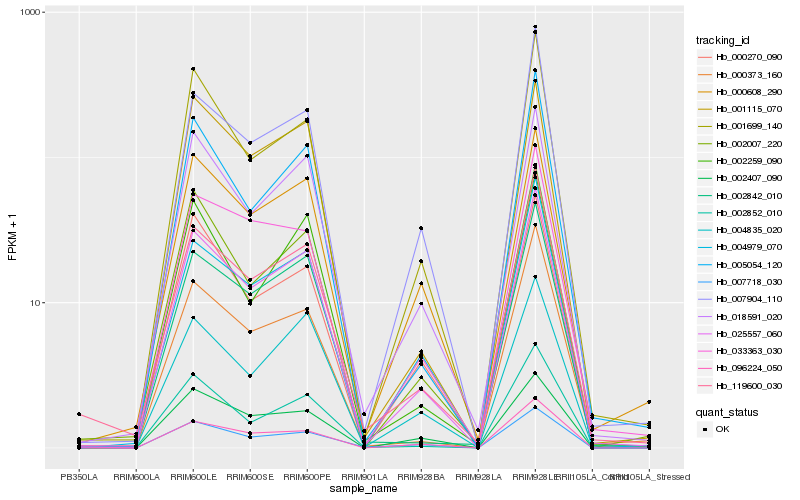
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025557_060 |
0.0 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_018591_020 |
0.0682464756 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_096224_050 |
0.0757819498 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 4 |
Hb_004979_070 |
0.0759445163 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 5 |
Hb_007904_110 |
0.076365652 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_001699_140 |
0.0764676945 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 7 |
Hb_119600_030 |
0.0775310061 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 8 |
Hb_005054_120 |
0.0792067981 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002842_010 |
0.0798664721 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 10 |
Hb_002852_010 |
0.0838694415 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_007718_030 |
0.087313154 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_004835_020 |
0.0873312163 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_000608_290 |
0.0888358609 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 14 |
Hb_000373_160 |
0.0907888319 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 15 |
Hb_002259_090 |
0.0911097226 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 16 |
Hb_033363_030 |
0.0922395614 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002007_220 |
0.0929497412 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002407_090 |
0.094565519 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 19 |
Hb_000270_090 |
0.0951300414 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 20 |
Hb_001115_070 |
0.0966789331 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |