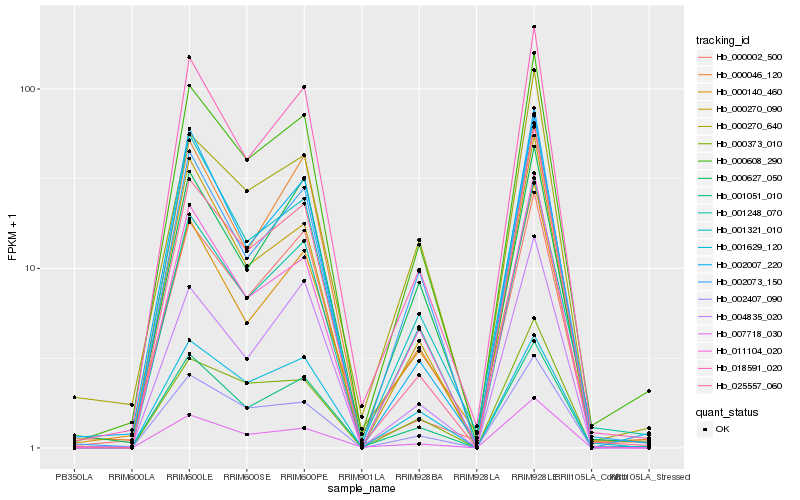
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_290 |
0.0 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 2 |
Hb_001051_010 |
0.0567082747 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 3 |
Hb_001248_070 |
0.0665609867 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 4 |
Hb_018591_020 |
0.0696405181 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_002407_090 |
0.0719841551 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 6 |
Hb_002073_150 |
0.0826056015 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 7 |
Hb_011104_020 |
0.0840668812 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 8 |
Hb_000140_460 |
0.0856001539 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 9 |
Hb_001321_010 |
0.0873619088 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 10 |
Hb_000373_010 |
0.087491786 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_025557_060 |
0.0888358609 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_007718_030 |
0.0919188295 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_000270_090 |
0.0947629533 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_000270_640 |
0.0950500361 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 15 |
Hb_001629_120 |
0.0983661067 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 16 |
Hb_000627_050 |
0.0986061582 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 17 |
Hb_000002_500 |
0.099347773 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004835_020 |
0.0993736848 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 19 |
Hb_002007_220 |
0.0995269869 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000046_120 |
0.1006719485 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |