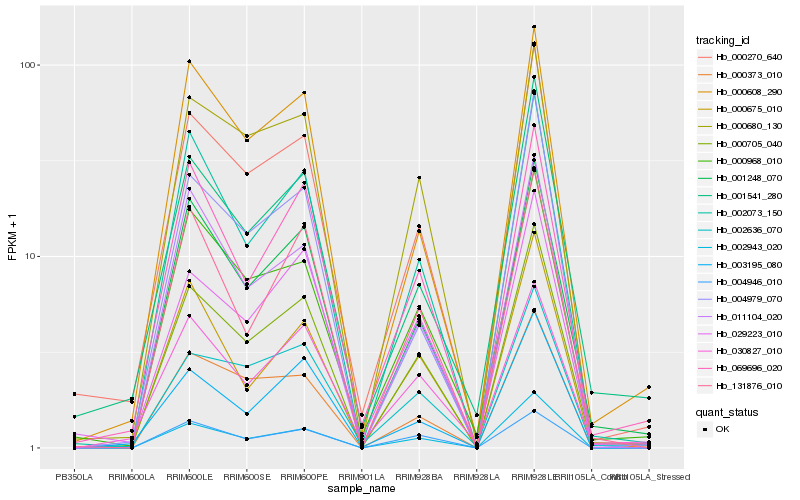
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000270_640 |
0.0736966471 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 3 |
Hb_002943_020 |
0.0743486252 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 4 |
Hb_001248_070 |
0.0747784326 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 5 |
Hb_000680_130 |
0.0792321414 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_002073_150 |
0.0836759705 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 7 |
Hb_029223_010 |
0.0879831155 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 8 |
Hb_002636_070 |
0.0893520149 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 9 |
Hb_000373_010 |
0.0911927749 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_069696_020 |
0.0980177094 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000968_010 |
0.0980686836 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011104_020 |
0.1026371101 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 13 |
Hb_003195_080 |
0.1037677565 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 14 |
Hb_131876_010 |
0.1048495881 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000608_290 |
0.1053946614 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 16 |
Hb_004979_070 |
0.1060718787 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 17 |
Hb_004946_010 |
0.10678829 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 18 |
Hb_001541_280 |
0.1086634301 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 19 |
Hb_000675_010 |
0.109662091 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 20 |
Hb_030827_010 |
0.1108951118 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |