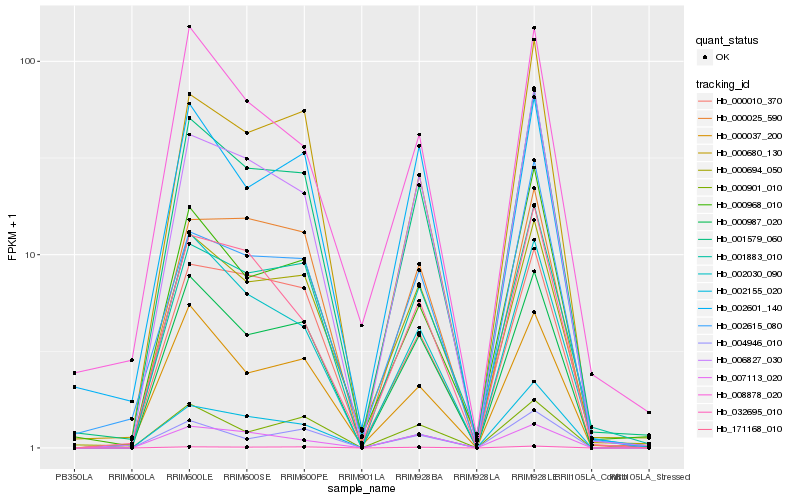
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001579_060 |
0.0 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 2 |
Hb_006827_030 |
0.0699622034 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 3 |
Hb_000694_050 |
0.0702800483 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 4 |
Hb_000987_020 |
0.0710525263 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 5 |
Hb_001883_010 |
0.0795132167 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 6 |
Hb_000680_130 |
0.0878519359 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 7 |
Hb_000968_010 |
0.0948239993 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_032695_010 |
0.0967919306 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 9 |
Hb_002030_090 |
0.1034549329 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_002155_020 |
0.1051772074 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 11 |
Hb_002615_080 |
0.1057555034 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 12 |
Hb_004946_010 |
0.1068904207 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 13 |
Hb_171168_010 |
0.1069699002 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_002601_140 |
0.1077362711 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 15 |
Hb_000037_200 |
0.1088573035 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000010_370 |
0.1091043251 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 17 |
Hb_000025_590 |
0.1119351867 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_007113_020 |
0.1159756319 |
- |
- |
- |
| 19 |
Hb_000901_010 |
0.1174263427 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 20 |
Hb_008878_020 |
0.1184913532 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |