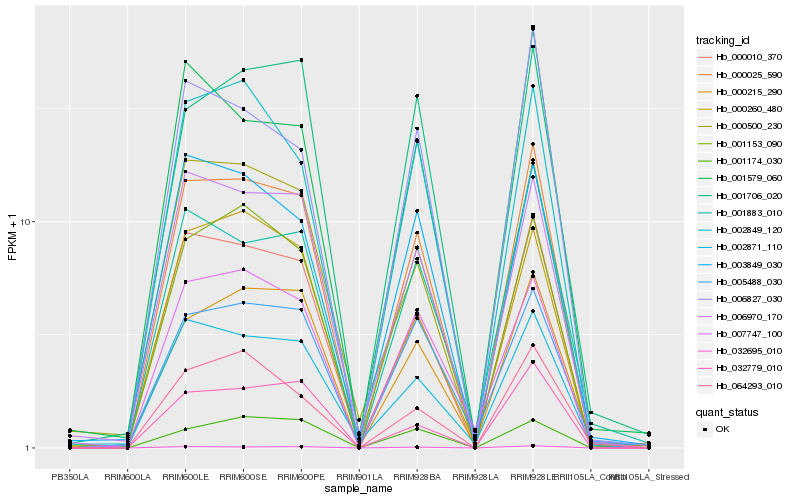
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_590 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000010_370 |
0.0616366105 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 3 |
Hb_032695_010 |
0.074300286 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 4 |
Hb_002871_110 |
0.0911313006 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 5 |
Hb_000215_290 |
0.0952884285 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006970_170 |
0.096611497 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_032779_010 |
0.0994929025 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_001883_010 |
0.0996421909 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 9 |
Hb_002849_120 |
0.1038655542 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 10 |
Hb_003849_030 |
0.1059649241 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 11 |
Hb_000500_230 |
0.1062349639 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 12 |
Hb_001153_090 |
0.1093860068 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 13 |
Hb_000260_480 |
0.1096499994 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 14 |
Hb_006827_030 |
0.1103936272 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 15 |
Hb_001706_020 |
0.1118555535 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 16 |
Hb_001579_060 |
0.1119351867 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 17 |
Hb_064293_010 |
0.1139753507 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_005488_030 |
0.1155569152 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 19 |
Hb_007747_100 |
0.1157956722 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 20 |
Hb_001174_030 |
0.1194734622 |
- |
- |
unnamed protein product [Vitis vinifera] |