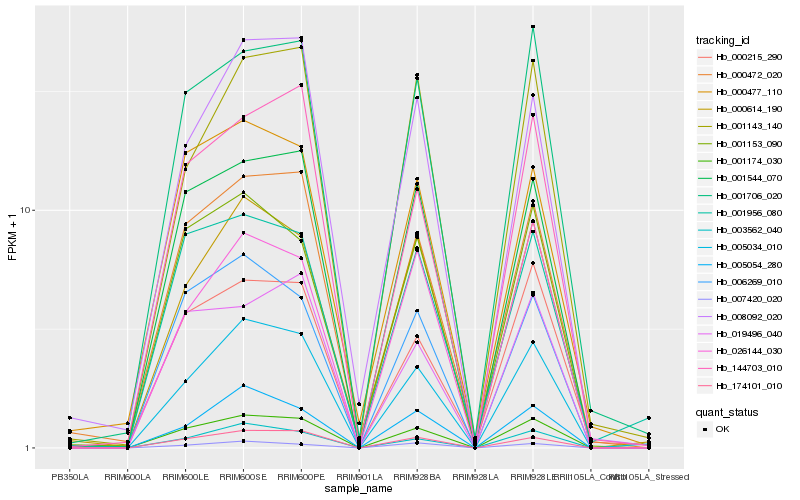
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001174_030 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_001544_070 |
0.0647250874 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 3 |
Hb_001706_020 |
0.0740886786 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 4 |
Hb_005034_010 |
0.0743980907 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_174101_010 |
0.0763948951 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 6 |
Hb_000472_020 |
0.0770914922 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 7 |
Hb_003562_040 |
0.0792933542 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 8 |
Hb_006269_010 |
0.0845355901 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 9 |
Hb_001153_090 |
0.0918796756 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 10 |
Hb_144703_010 |
0.0940676048 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 11 |
Hb_019496_040 |
0.0942744568 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 12 |
Hb_000614_190 |
0.0965108765 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 13 |
Hb_000215_290 |
0.0965744301 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_026144_030 |
0.1019114166 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 15 |
Hb_005054_280 |
0.1021801213 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 16 |
Hb_001956_080 |
0.1109698767 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 17 |
Hb_000477_110 |
0.1117733102 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 18 |
Hb_007420_020 |
0.1132127978 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_001143_140 |
0.1148336861 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_008092_020 |
0.1150039436 |
- |
- |
transcription factor, putative [Ricinus communis] |