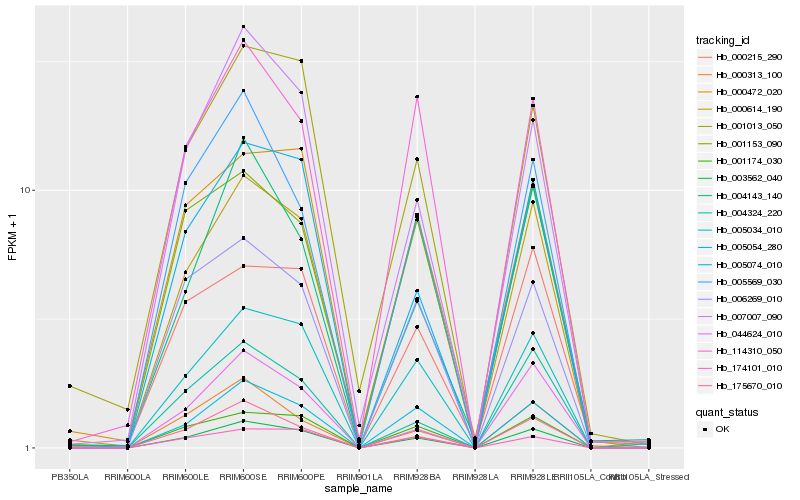
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003562_040 |
0.0 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 2 |
Hb_005034_010 |
0.0748185261 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_001174_030 |
0.0792933542 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_005054_280 |
0.0835847114 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 5 |
Hb_005074_010 |
0.085326374 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 6 |
Hb_175670_010 |
0.0884820609 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_007007_090 |
0.1012961732 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 8 |
Hb_006269_010 |
0.1043306286 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 9 |
Hb_000614_190 |
0.1101450982 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 10 |
Hb_004324_220 |
0.1106820523 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 11 |
Hb_005569_030 |
0.1119651133 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 12 |
Hb_174101_010 |
0.1130610171 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_000313_100 |
0.1136447091 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 14 |
Hb_000472_020 |
0.1167754856 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 15 |
Hb_114310_050 |
0.1176376107 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 16 |
Hb_001013_050 |
0.1192408902 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_044624_010 |
0.1199607686 |
- |
- |
- |
| 18 |
Hb_001153_090 |
0.1219889687 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 19 |
Hb_000215_290 |
0.1226679717 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004143_140 |
0.1258322363 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |