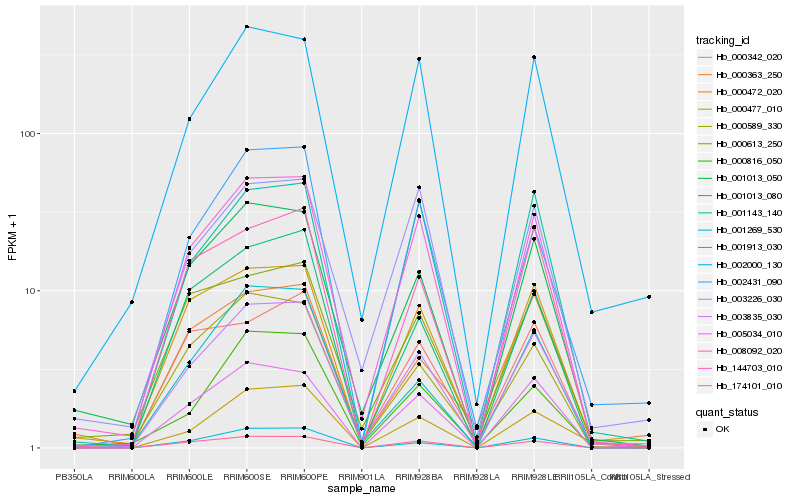
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003835_030 |
0.0 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 2 |
Hb_008092_020 |
0.0665084639 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001013_050 |
0.0767807617 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_002000_130 |
0.0951091191 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 5 |
Hb_000589_330 |
0.0978189253 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_005034_010 |
0.0979448739 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000472_020 |
0.0998289282 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 8 |
Hb_002431_090 |
0.1084051851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001913_030 |
0.1084570124 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 10 |
Hb_000477_010 |
0.1102286733 |
- |
- |
PREDICTED: potassium channel AKT1-like [Jatropha curcas] |
| 11 |
Hb_144703_010 |
0.1102682595 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 12 |
Hb_174101_010 |
0.1110233424 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_001269_530 |
0.1122695898 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_000613_250 |
0.1136229324 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 15 |
Hb_001013_080 |
0.1204579851 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 16 |
Hb_000363_250 |
0.1218322256 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 17 |
Hb_001143_140 |
0.1220702576 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000816_050 |
0.123714498 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_003226_030 |
0.12825541 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 20 |
Hb_000342_020 |
0.1298110894 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |