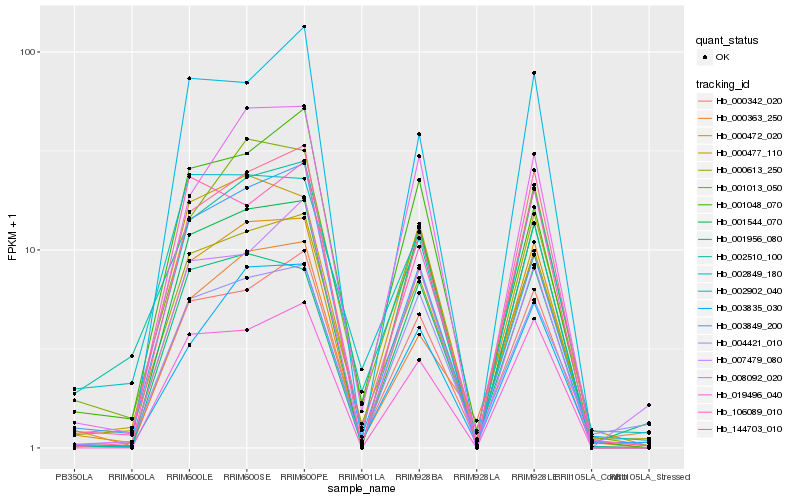
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_250 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 2 |
Hb_000342_020 |
0.0745010884 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 3 |
Hb_000472_020 |
0.0762467168 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 4 |
Hb_004421_010 |
0.092774387 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 5 |
Hb_000477_110 |
0.0931844973 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 6 |
Hb_001013_050 |
0.1025477634 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_144703_010 |
0.1052334846 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 8 |
Hb_002849_180 |
0.1053516286 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 9 |
Hb_002510_100 |
0.1075413651 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 10 |
Hb_003849_200 |
0.1076233862 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_001048_070 |
0.1094673025 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 12 |
Hb_106089_010 |
0.1115378091 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 13 |
Hb_003835_030 |
0.1136229324 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 14 |
Hb_001544_070 |
0.1171335932 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 15 |
Hb_019496_040 |
0.1179253377 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 16 |
Hb_002902_040 |
0.1191761603 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 17 |
Hb_007479_080 |
0.1204179717 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000363_250 |
0.120623493 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 19 |
Hb_008092_020 |
0.121805819 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001956_080 |
0.1218432112 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |