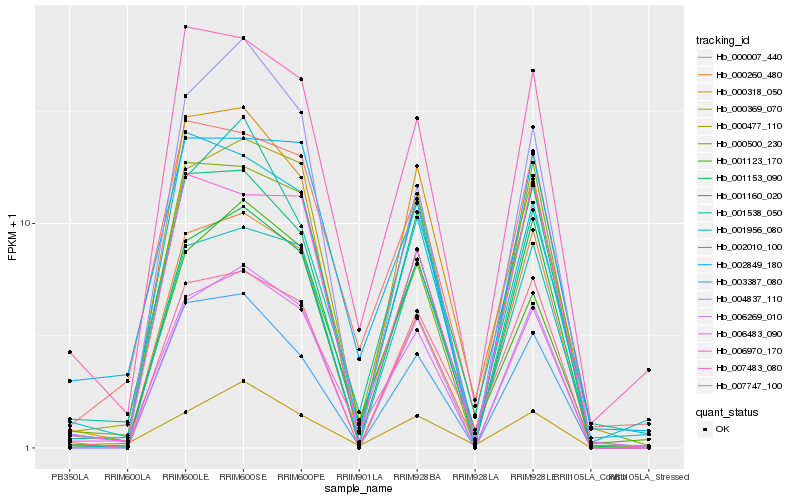
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006483_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 2 |
Hb_000477_110 |
0.0758489064 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 3 |
Hb_002849_180 |
0.0988684891 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 4 |
Hb_006269_010 |
0.1016549195 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 5 |
Hb_001153_090 |
0.1032179003 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 6 |
Hb_000007_440 |
0.1038897727 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 7 |
Hb_007483_080 |
0.1044997021 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 8 |
Hb_001160_020 |
0.1137902221 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 9 |
Hb_003387_080 |
0.1170157724 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000500_230 |
0.1173518365 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 11 |
Hb_000260_480 |
0.1202059305 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 12 |
Hb_001123_170 |
0.1202393082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001538_050 |
0.1233497924 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 14 |
Hb_000318_050 |
0.1238780224 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000369_070 |
0.1255119554 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 16 |
Hb_002010_100 |
0.1261435211 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 17 |
Hb_006970_170 |
0.1271286278 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_007747_100 |
0.1273802676 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 19 |
Hb_001956_080 |
0.1291775013 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 20 |
Hb_004837_110 |
0.1297860014 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |