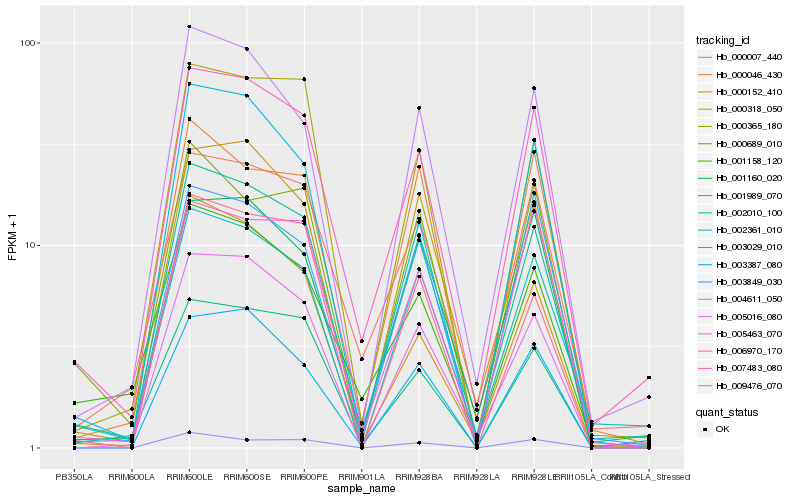
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002010_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 2 |
Hb_007483_080 |
0.0636104697 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 3 |
Hb_002361_010 |
0.0736507834 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_005016_080 |
0.0782531477 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 5 |
Hb_005463_070 |
0.0793445781 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_000318_050 |
0.0921237069 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000365_180 |
0.0988419811 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 8 |
Hb_001989_070 |
0.1002727625 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 9 |
Hb_000007_440 |
0.1026305567 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 10 |
Hb_003387_080 |
0.1115388185 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001158_120 |
0.1155518482 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003029_010 |
0.1156181956 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009476_070 |
0.1162604582 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 14 |
Hb_000046_430 |
0.1163498015 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 15 |
Hb_000152_410 |
0.1164925152 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 16 |
Hb_000689_010 |
0.1180635931 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_001160_020 |
0.1190691672 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 18 |
Hb_004611_050 |
0.1197169134 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 19 |
Hb_006970_170 |
0.1213461802 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_003849_030 |
0.121354711 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |