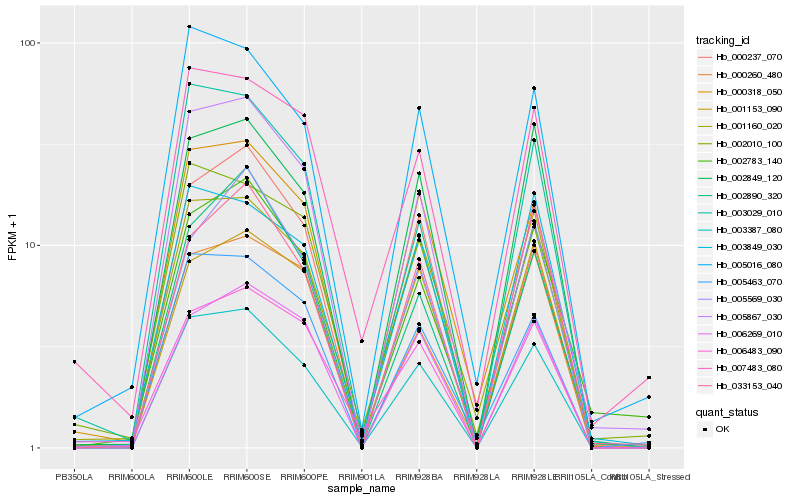
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003387_080 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000318_050 |
0.0863230648 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005016_080 |
0.0967791218 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 4 |
Hb_002849_120 |
0.0967846898 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_005463_070 |
0.097370382 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_001160_020 |
0.0982327548 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 7 |
Hb_006269_010 |
0.1008009142 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 8 |
Hb_000237_070 |
0.1070677693 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 9 |
Hb_003029_010 |
0.1072463683 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003849_030 |
0.1090396518 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 11 |
Hb_002010_100 |
0.1115388185 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 12 |
Hb_005867_030 |
0.1161453859 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 13 |
Hb_006483_090 |
0.1170157724 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 14 |
Hb_002783_140 |
0.1172908132 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 15 |
Hb_000260_480 |
0.1194211878 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 16 |
Hb_001153_090 |
0.1205757057 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 17 |
Hb_002890_320 |
0.1210588435 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 18 |
Hb_033153_040 |
0.1214786037 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_007483_080 |
0.1219209948 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 20 |
Hb_005569_030 |
0.1232407278 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |