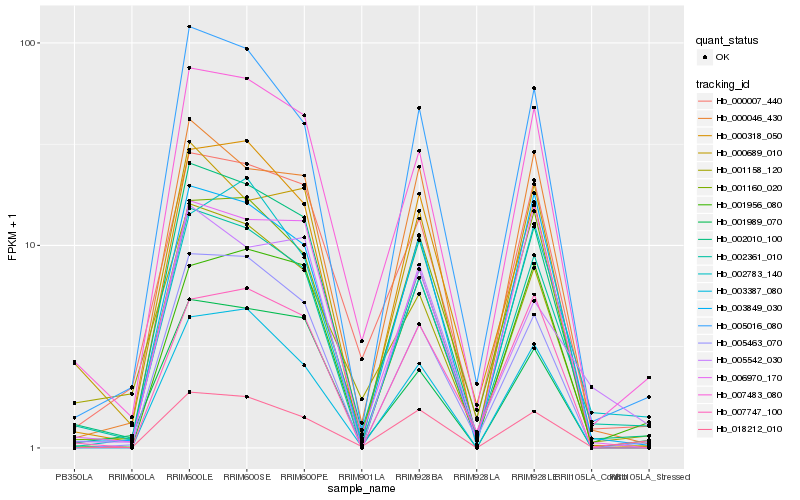
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002361_010 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002010_100 |
0.0736507834 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 3 |
Hb_005016_080 |
0.0810749672 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 4 |
Hb_007483_080 |
0.0931899241 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 5 |
Hb_005463_070 |
0.1084108182 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_000318_050 |
0.1085158158 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 7 |
Hb_018212_010 |
0.1110855859 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 8 |
Hb_003849_030 |
0.1118549529 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 9 |
Hb_000046_430 |
0.1175636803 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 10 |
Hb_001160_020 |
0.1198315524 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 11 |
Hb_000007_440 |
0.1244571319 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 12 |
Hb_000689_010 |
0.125865706 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_003387_080 |
0.1299579088 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001956_080 |
0.1338788343 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 15 |
Hb_002783_140 |
0.1347691883 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 16 |
Hb_006970_170 |
0.1369639747 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001158_120 |
0.1378197455 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007747_100 |
0.1378921103 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 19 |
Hb_001989_070 |
0.1381487067 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 20 |
Hb_005542_030 |
0.1396924008 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 isoform X2 [Jatropha curcas] |