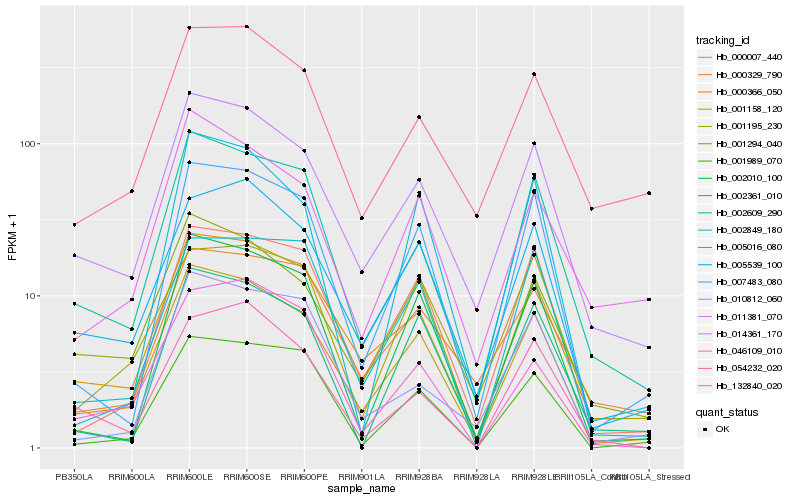
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011381_070 |
0.0775808482 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_000366_050 |
0.080674697 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_001294_040 |
0.0932778186 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 5 |
Hb_002609_290 |
0.099867162 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 6 |
Hb_005539_100 |
0.1021903688 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 7 |
Hb_007483_080 |
0.1033903233 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 8 |
Hb_000007_440 |
0.1066959105 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_002010_100 |
0.1155518482 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 10 |
Hb_014361_170 |
0.1275093614 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 11 |
Hb_001989_070 |
0.1290403298 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 12 |
Hb_002849_180 |
0.1302186628 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 13 |
Hb_005016_080 |
0.1369195159 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 14 |
Hb_000329_790 |
0.1375713231 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 15 |
Hb_002361_010 |
0.1378197455 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_132840_020 |
0.139859322 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 17 |
Hb_001195_230 |
0.1449269844 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 18 |
Hb_010812_060 |
0.148971833 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 19 |
Hb_046109_010 |
0.149785794 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 20 |
Hb_054232_020 |
0.1499828268 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |