| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
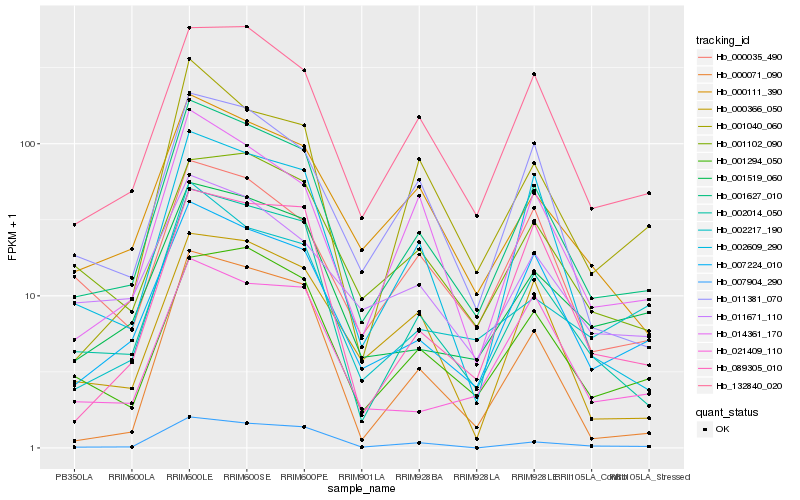
Hb_001627_010 |
0.0 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 2 |
Hb_002014_050 |
0.0770318854 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 3 |
Hb_007224_010 |
0.0971380282 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 4 |
Hb_132840_020 |
0.1009724938 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 5 |
Hb_001294_050 |
0.1078316096 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 6 |
Hb_001519_060 |
0.1093788896 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000111_390 |
0.1126924938 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 8 |
Hb_014361_170 |
0.1133978029 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 9 |
Hb_011381_070 |
0.1250028011 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 10 |
Hb_002609_290 |
0.1252907977 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 11 |
Hb_007904_290 |
0.1258262881 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 12 |
Hb_001040_060 |
0.1324469899 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 13 |
Hb_001102_090 |
0.1333777583 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 14 |
Hb_000035_490 |
0.1343020248 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 15 |
Hb_002217_190 |
0.1364278586 |
- |
- |
invertase [Hevea brasiliensis] |
| 16 |
Hb_000071_090 |
0.1376948741 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 17 |
Hb_000366_050 |
0.1385993833 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_011671_110 |
0.1388029426 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 19 |
Hb_021409_110 |
0.1401438719 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_089305_010 |
0.1401439138 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |