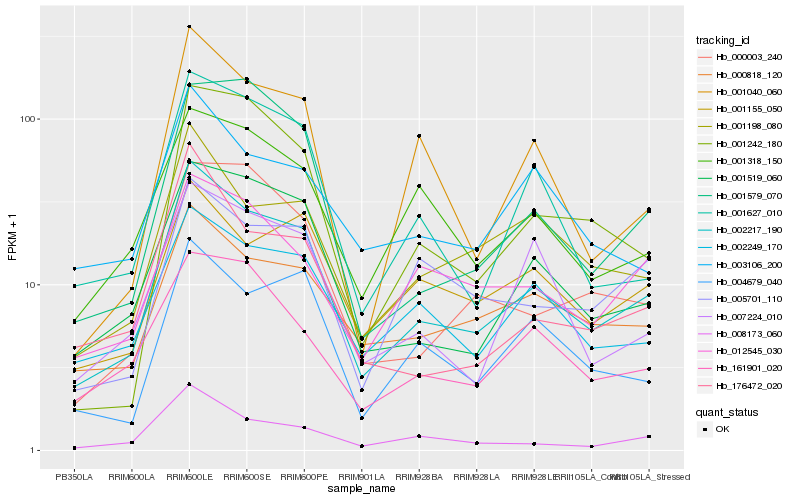
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_190 |
0.0 |
- |
- |
invertase [Hevea brasiliensis] |
| 2 |
Hb_001519_060 |
0.1158708327 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_008173_060 |
0.1239786554 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_000818_120 |
0.1363388912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001627_010 |
0.1364278586 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 6 |
Hb_176472_020 |
0.1378112179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001198_080 |
0.1385673534 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004679_040 |
0.1396207049 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 9 |
Hb_002249_170 |
0.1404481159 |
- |
- |
copine, putative [Ricinus communis] |
| 10 |
Hb_001040_060 |
0.144087287 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 11 |
Hb_001318_150 |
0.1495227944 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 12 |
Hb_007224_010 |
0.1500008043 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 13 |
Hb_001155_050 |
0.1503863423 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 14 |
Hb_161901_020 |
0.1505738863 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 15 |
Hb_001579_070 |
0.153358358 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 16 |
Hb_000003_240 |
0.1537605512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001242_180 |
0.1546567828 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 18 |
Hb_003106_200 |
0.1553351076 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 19 |
Hb_012545_030 |
0.1566923578 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 20 |
Hb_005701_110 |
0.1567466289 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |