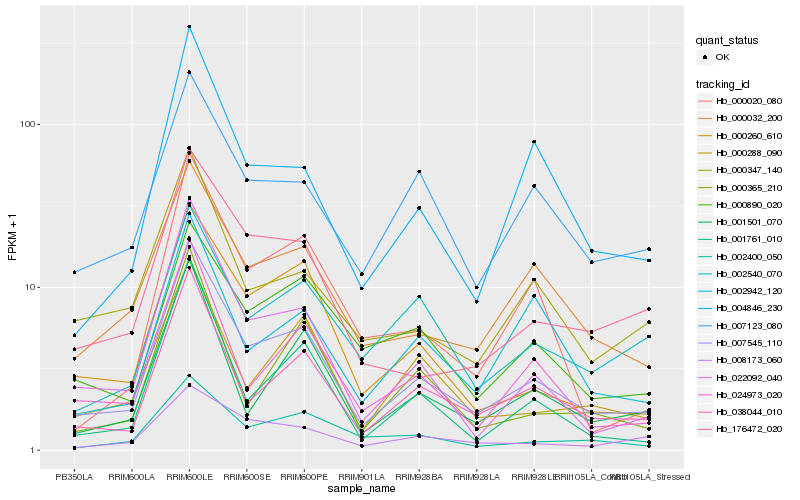
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007545_110 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_038044_010 |
0.1055797382 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 3 |
Hb_022092_040 |
0.1176766611 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 4 |
Hb_000260_610 |
0.1273478104 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 5 |
Hb_024973_020 |
0.1274489494 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 6 |
Hb_000032_200 |
0.1333531861 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_176472_020 |
0.1392249766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001501_070 |
0.1413182159 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 9 |
Hb_001761_010 |
0.1415924404 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 10 |
Hb_002400_050 |
0.1426616595 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004846_230 |
0.1430052578 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000347_140 |
0.1454624003 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_007123_080 |
0.1487255799 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 14 |
Hb_000288_090 |
0.149430139 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000890_020 |
0.151222633 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_000020_080 |
0.1526945095 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 17 |
Hb_008173_060 |
0.1566662597 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 18 |
Hb_002540_070 |
0.1586656484 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 19 |
Hb_000365_210 |
0.1588007453 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 20 |
Hb_002942_120 |
0.1590982224 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |