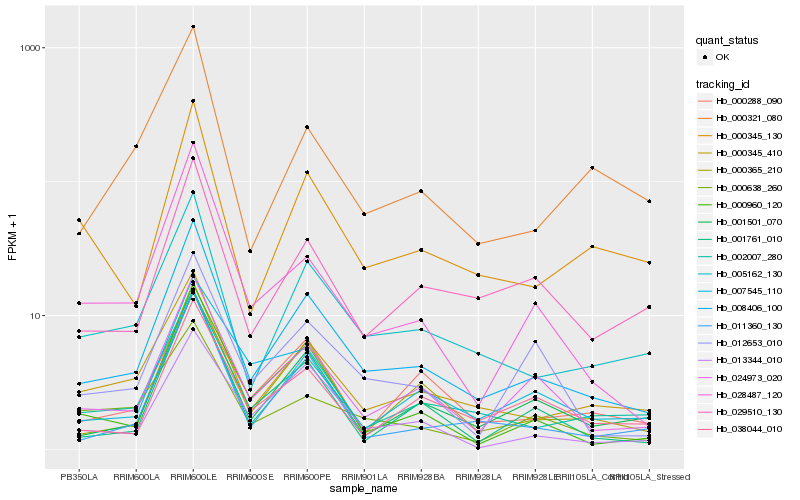
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_100 |
0.0 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005162_130 |
0.1085017252 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002007_280 |
0.1136808862 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 4 |
Hb_000345_410 |
0.1157856073 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 5 |
Hb_024973_020 |
0.1160392086 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 6 |
Hb_000365_210 |
0.1222547374 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 7 |
Hb_001501_070 |
0.1253423733 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 8 |
Hb_011360_130 |
0.1269238675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000960_120 |
0.1296574731 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 10 |
Hb_001761_010 |
0.1387922243 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 11 |
Hb_028487_120 |
0.1393301549 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 12 |
Hb_013344_010 |
0.1461853057 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 13 |
Hb_029510_130 |
0.1490878687 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 14 |
Hb_000638_260 |
0.1498072804 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 15 |
Hb_038044_010 |
0.1504612903 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 16 |
Hb_000345_130 |
0.1530721202 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_012653_010 |
0.1559252792 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 18 |
Hb_000321_080 |
0.1595037917 |
- |
- |
histone H4 [Zea mays] |
| 19 |
Hb_000288_090 |
0.1626282471 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_007545_110 |
0.16293967 |
- |
- |
catalytic, putative [Ricinus communis] |