| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
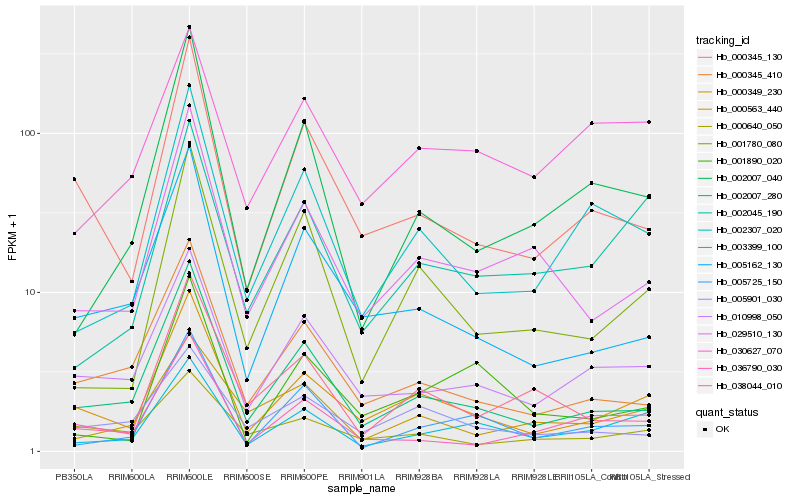
Hb_000349_230 |
0.0 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 2 |
Hb_002007_280 |
0.1210601185 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 3 |
Hb_000345_130 |
0.1211933089 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_010998_050 |
0.1293251028 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005162_130 |
0.1364797828 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000345_410 |
0.1445154428 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 7 |
Hb_029510_130 |
0.1488177594 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 8 |
Hb_002307_020 |
0.1637560804 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 9 |
Hb_001780_080 |
0.1639215611 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 10 |
Hb_000640_050 |
0.1651774391 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 11 |
Hb_005901_030 |
0.1676096596 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003399_100 |
0.1680018852 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 13 |
Hb_036790_030 |
0.174145334 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 14 |
Hb_005725_150 |
0.1816107033 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002045_190 |
0.1873318564 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 16 |
Hb_001890_020 |
0.1890479412 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 17 |
Hb_038044_010 |
0.1907121135 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 18 |
Hb_000563_440 |
0.1934666282 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002007_040 |
0.1939651724 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_030627_070 |
0.1942572032 |
- |
- |
histone H4 [Zea mays] |