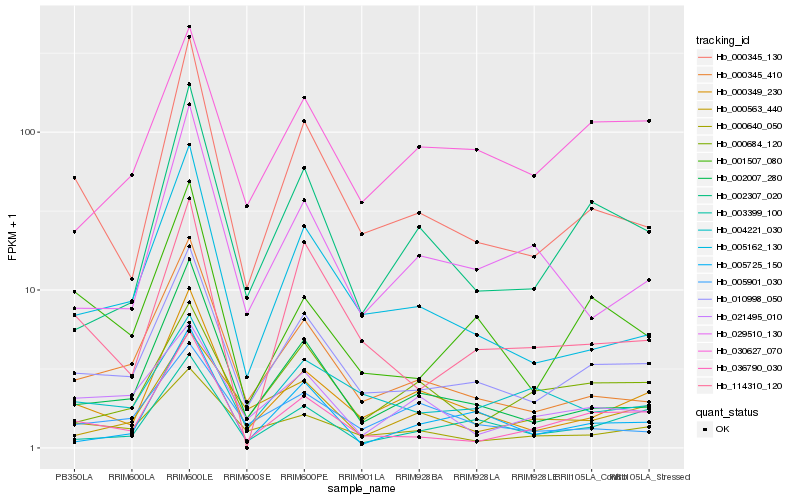
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010998_050 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 2 |
Hb_036790_030 |
0.123479694 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 3 |
Hb_000345_410 |
0.1256828049 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 4 |
Hb_002007_280 |
0.127824732 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 5 |
Hb_000349_230 |
0.1293251028 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 6 |
Hb_000345_130 |
0.1306046896 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_005162_130 |
0.1377502008 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000640_050 |
0.1377983707 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 9 |
Hb_030627_070 |
0.147235063 |
- |
- |
histone H4 [Zea mays] |
| 10 |
Hb_003399_100 |
0.1514734672 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 11 |
Hb_001507_080 |
0.1519401658 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 12 |
Hb_002307_020 |
0.1562497093 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 13 |
Hb_004221_030 |
0.1607273084 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005901_030 |
0.1629485361 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_114310_120 |
0.1634724046 |
- |
- |
PREDICTED: 50S ribosomal protein L31, chloroplastic [Eucalyptus grandis] |
| 16 |
Hb_021495_010 |
0.1659939003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000563_440 |
0.1686255696 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000684_120 |
0.1693950397 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005725_150 |
0.1712071736 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_029510_130 |
0.1717465502 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |