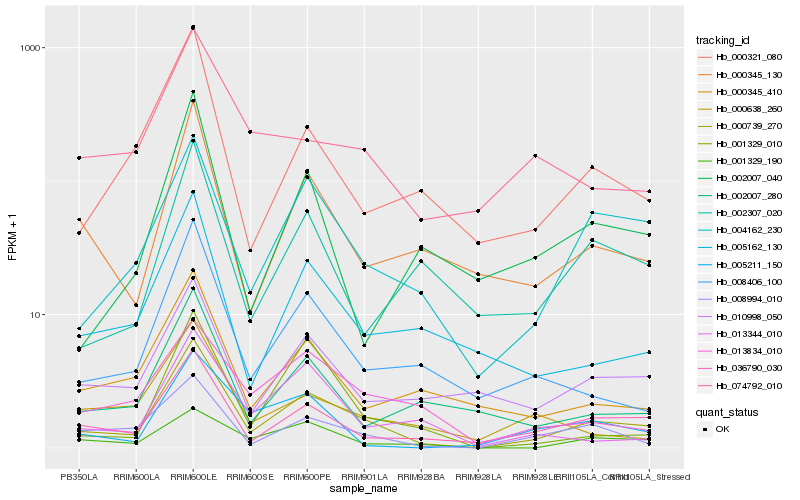
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_270 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 2 |
Hb_036790_030 |
0.1707313436 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 3 |
Hb_004162_230 |
0.1824435547 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 4 |
Hb_008994_010 |
0.1897296286 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_000345_130 |
0.1924812432 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_008406_100 |
0.1991792614 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_010998_050 |
0.2026989569 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005162_130 |
0.2032337118 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000321_080 |
0.2040894742 |
- |
- |
histone H4 [Zea mays] |
| 10 |
Hb_001329_190 |
0.2046636793 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 11 |
Hb_000345_410 |
0.2095070948 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 12 |
Hb_002007_280 |
0.2131808547 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 13 |
Hb_013834_010 |
0.2216597586 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 14 |
Hb_013344_010 |
0.2219055601 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 15 |
Hb_002307_020 |
0.2221713646 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 16 |
Hb_005211_150 |
0.223828039 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38) isoform X1 [Jatropha curcas] |
| 17 |
Hb_001329_010 |
0.2244566983 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000638_260 |
0.2291317114 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 19 |
Hb_002007_040 |
0.2295906577 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_074792_010 |
0.2335709125 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |