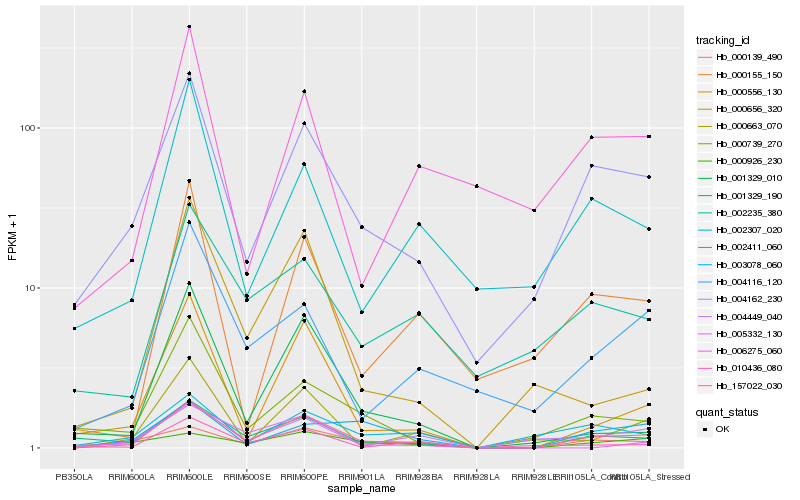
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_157022_030 |
0.0 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 2 |
Hb_004162_230 |
0.1627976611 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 3 |
Hb_000139_490 |
0.1723691185 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 4 |
Hb_001329_190 |
0.1980832936 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_000556_130 |
0.2091972928 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 6 |
Hb_000663_070 |
0.2234761194 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 7 |
Hb_004449_040 |
0.2264698414 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 8 |
Hb_004116_120 |
0.2315874151 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 9 |
Hb_002411_060 |
0.2322412165 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 10 |
Hb_010436_080 |
0.235143716 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 11 |
Hb_000739_270 |
0.2376944353 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 12 |
Hb_000155_150 |
0.2378777496 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 13 |
Hb_003078_060 |
0.238149452 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 14 |
Hb_002235_380 |
0.2399262089 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 15 |
Hb_002307_020 |
0.240245332 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 16 |
Hb_001329_010 |
0.246308597 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000926_230 |
0.2466234579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000656_320 |
0.2559658203 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 19 |
Hb_006275_060 |
0.2570846997 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 20 |
Hb_005332_130 |
0.2611676695 |
- |
- |
Mavicyanin, putative [Ricinus communis] |