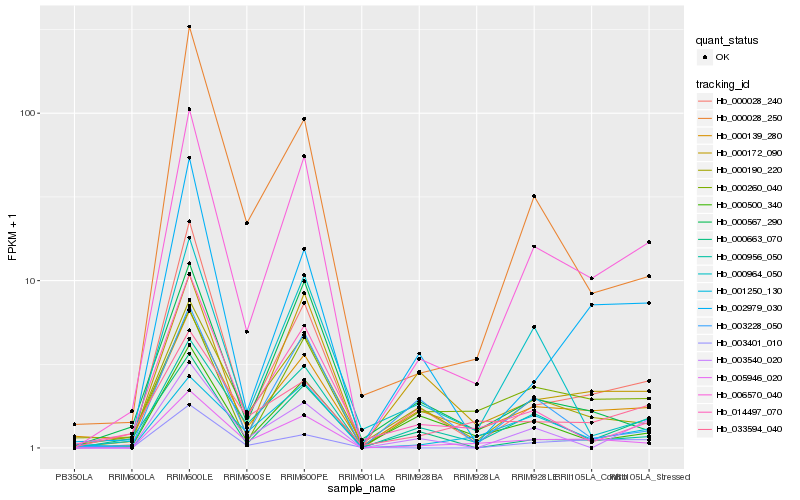
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_040 |
0.0 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 2 |
Hb_005946_020 |
0.1384956913 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |
| 3 |
Hb_000028_240 |
0.1612987533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000139_280 |
0.1684351133 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000190_220 |
0.1794586587 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 6 |
Hb_000260_040 |
0.1806825739 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 7 |
Hb_000956_050 |
0.1811359221 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003401_010 |
0.1834187327 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 9 |
Hb_000663_070 |
0.1860387887 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 10 |
Hb_000172_090 |
0.1869547026 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 11 |
Hb_002979_030 |
0.1881434033 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003540_020 |
0.1949599723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001250_130 |
0.1950710624 |
- |
- |
hypothetical protein POPTR_0008s19280g [Populus trichocarpa] |
| 14 |
Hb_000028_250 |
0.1967182201 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 15 |
Hb_014497_070 |
0.2001193843 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 16 |
Hb_000500_340 |
0.2042590118 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 17 |
Hb_000964_050 |
0.2044147998 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 18 |
Hb_033594_040 |
0.2045833462 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_003228_050 |
0.2046378144 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 20 |
Hb_000567_290 |
0.2060018109 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |