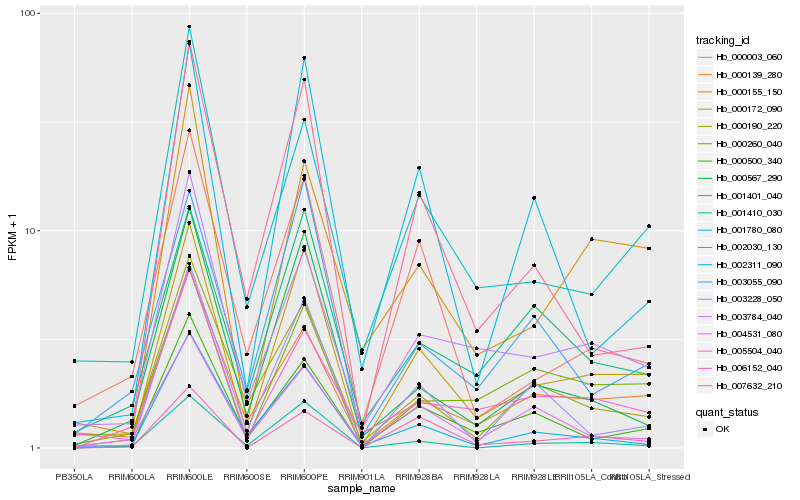
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 2 |
Hb_000500_340 |
0.1444110348 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 3 |
Hb_000139_280 |
0.1545235804 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000260_040 |
0.1551127066 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 5 |
Hb_003228_050 |
0.1582135478 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 6 |
Hb_007632_210 |
0.1589872806 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 7 |
Hb_000155_150 |
0.1609435689 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 8 |
Hb_001410_030 |
0.1617300108 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 9 |
Hb_002030_130 |
0.1617579178 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 10 |
Hb_000567_290 |
0.1620861528 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 11 |
Hb_006152_040 |
0.1653697293 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105645062 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003055_090 |
0.1655108314 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 13 |
Hb_003784_040 |
0.1665289866 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 14 |
Hb_001401_040 |
0.1669943849 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 15 |
Hb_000190_220 |
0.1685651807 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 16 |
Hb_000003_060 |
0.1715121449 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 17 |
Hb_005504_040 |
0.1729537262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004531_080 |
0.1745947524 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 19 |
Hb_002311_090 |
0.1750486633 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 20 |
Hb_001780_080 |
0.1762894967 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |