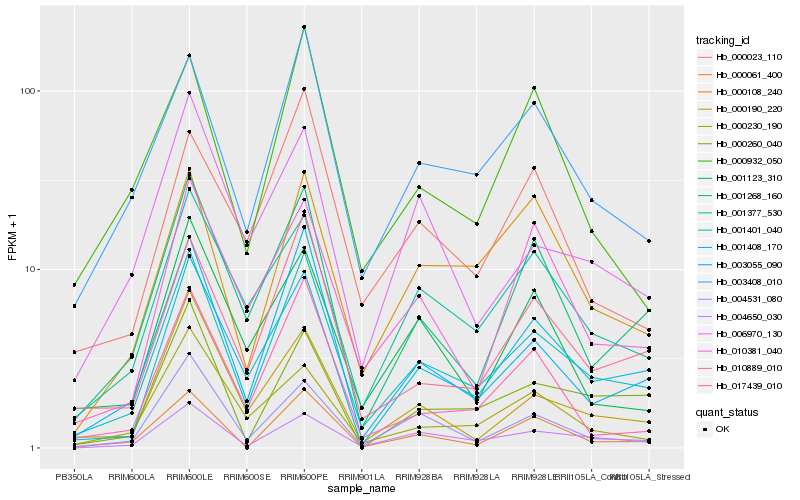
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001401_040 |
0.0 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 2 |
Hb_003055_090 |
0.0995790674 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 3 |
Hb_001408_170 |
0.1189009926 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 4 |
Hb_003408_010 |
0.1191319236 |
- |
- |
fructokinase [Manihot esculenta] |
| 5 |
Hb_004650_030 |
0.1196193574 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 6 |
Hb_017439_010 |
0.1201445156 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 7 |
Hb_001268_160 |
0.1302099601 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 8 |
Hb_001377_530 |
0.1388408038 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_004531_080 |
0.1393809222 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 10 |
Hb_000061_400 |
0.1462221629 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_010889_010 |
0.1464079297 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 12 |
Hb_000260_040 |
0.1479005768 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 13 |
Hb_000190_220 |
0.1525327959 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 14 |
Hb_000108_240 |
0.1535444059 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 15 |
Hb_010381_040 |
0.1596678856 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 16 |
Hb_000230_190 |
0.1614430391 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006970_130 |
0.1614857551 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 18 |
Hb_001123_310 |
0.1622312769 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 19 |
Hb_000023_110 |
0.1627218245 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 20 |
Hb_000932_050 |
0.1629777018 |
- |
- |
annexin [Manihot esculenta] |