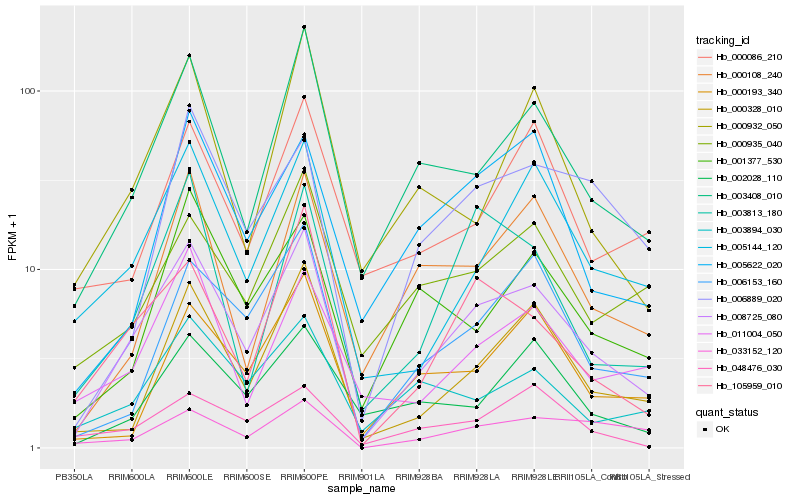
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_080 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 2 |
Hb_003408_010 |
0.1393273318 |
- |
- |
fructokinase [Manihot esculenta] |
| 3 |
Hb_000328_010 |
0.1612796883 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 4 |
Hb_105959_010 |
0.1629840625 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 5 |
Hb_005144_120 |
0.1634969148 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 6 |
Hb_006153_160 |
0.1704675772 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X4 [Jatropha curcas] |
| 7 |
Hb_000108_240 |
0.1782077724 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 8 |
Hb_003894_030 |
0.1818916402 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 9 |
Hb_011004_050 |
0.1820995542 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 10 |
Hb_000935_040 |
0.1833776184 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_003813_180 |
0.1842941197 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 12 |
Hb_005622_020 |
0.1855110144 |
- |
- |
PREDICTED: DCC family protein At1g52590, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000193_340 |
0.1857100097 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001377_530 |
0.1865683968 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_002028_110 |
0.187038342 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 16 |
Hb_000932_050 |
0.1876866794 |
- |
- |
annexin [Manihot esculenta] |
| 17 |
Hb_048476_030 |
0.193970055 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |
| 18 |
Hb_006889_020 |
0.1953470744 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 19 |
Hb_000086_210 |
0.1959577124 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_033152_120 |
0.1970944929 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |