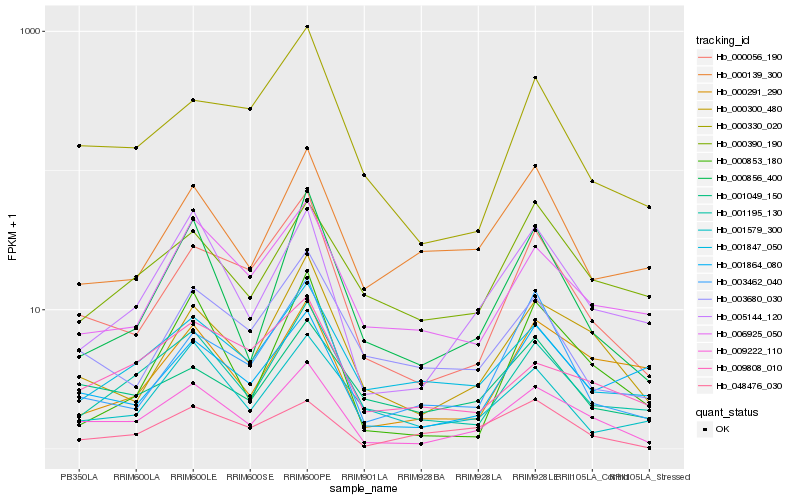
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009222_110 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 2 |
Hb_000300_480 |
0.1348769865 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 3 |
Hb_005144_120 |
0.1402035674 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 4 |
Hb_000056_190 |
0.1445983746 |
- |
- |
hypothetical protein [Ricinus communis] |
| 5 |
Hb_001195_130 |
0.1526902878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000856_400 |
0.1657620882 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 7 |
Hb_006925_050 |
0.1730033903 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 8 |
Hb_001847_050 |
0.1732719805 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 9 |
Hb_001049_150 |
0.1778341318 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 10 |
Hb_000330_020 |
0.1797098057 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 11 |
Hb_009808_010 |
0.1851351596 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_000291_290 |
0.1852881394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003680_030 |
0.1869890771 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000390_190 |
0.188635568 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_001864_080 |
0.1889767076 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 16 |
Hb_003462_040 |
0.1900102557 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_048476_030 |
0.1909302218 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |
| 18 |
Hb_001579_300 |
0.1923574737 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 19 |
Hb_000139_300 |
0.1924135292 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 20 |
Hb_000853_180 |
0.1926914171 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |