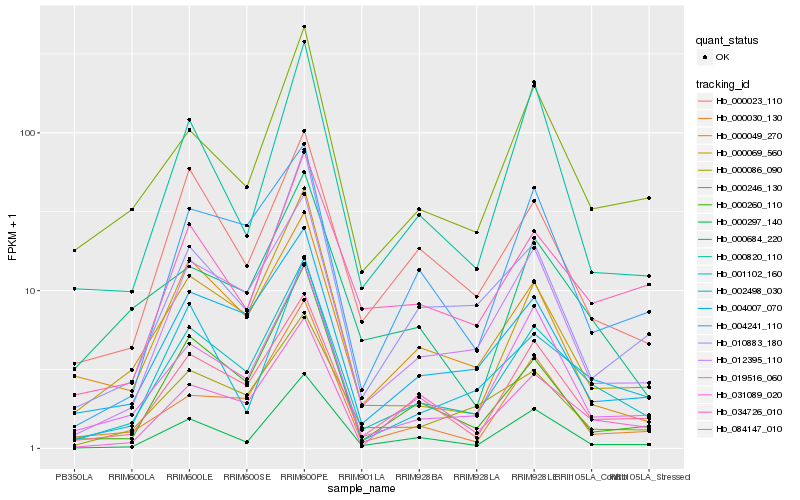
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_160 |
0.0 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 2 |
Hb_084147_010 |
0.1027751874 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_012395_110 |
0.1083918157 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 4 |
Hb_004007_070 |
0.1210789987 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 5 |
Hb_019516_060 |
0.1217469924 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 6 |
Hb_000086_090 |
0.1247859468 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 7 |
Hb_000246_130 |
0.1274140677 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 8 |
Hb_000069_560 |
0.1274346141 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 9 |
Hb_000260_110 |
0.1336222476 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_031089_020 |
0.1369325644 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 11 |
Hb_002498_030 |
0.1403187087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010883_180 |
0.1419535165 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 13 |
Hb_000297_140 |
0.1428890706 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 14 |
Hb_034726_010 |
0.1434350251 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_000023_110 |
0.1456557156 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 16 |
Hb_004241_110 |
0.1469156122 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 17 |
Hb_000030_130 |
0.1515492896 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000820_110 |
0.1541132131 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000049_270 |
0.1547881754 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000684_220 |
0.1558633622 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |