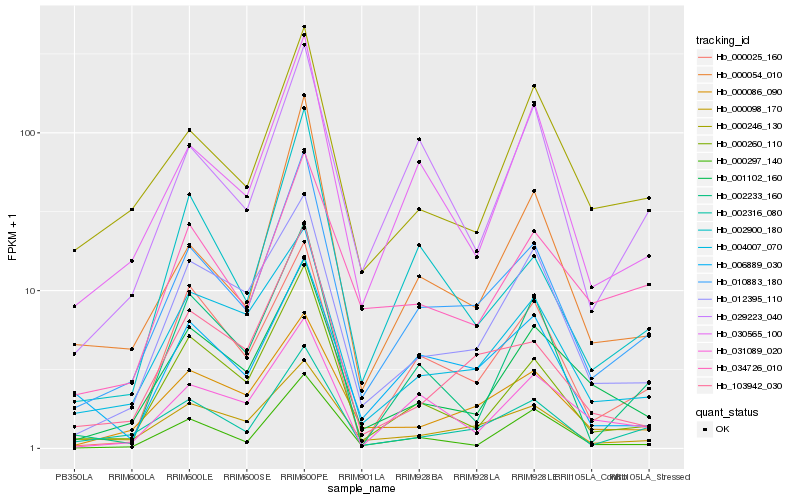
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_180 |
0.0 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 2 |
Hb_002316_080 |
0.1103164308 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 3 |
Hb_030565_100 |
0.119831 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000260_110 |
0.1225961996 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_103942_030 |
0.1318055871 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_000054_010 |
0.1330153631 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000086_090 |
0.1337205724 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_000098_170 |
0.1361971142 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 9 |
Hb_004007_070 |
0.1381670202 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 10 |
Hb_012395_110 |
0.1415784265 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 11 |
Hb_001102_160 |
0.1419535165 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 12 |
Hb_000297_140 |
0.1453586906 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 13 |
Hb_002900_180 |
0.1458368532 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 14 |
Hb_000246_130 |
0.1469942447 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 15 |
Hb_029223_040 |
0.1516746628 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 16 |
Hb_000025_160 |
0.1519188825 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002233_160 |
0.1561743756 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 18 |
Hb_006889_030 |
0.1563197767 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 19 |
Hb_031089_020 |
0.1565464755 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 20 |
Hb_034726_010 |
0.1574629528 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |