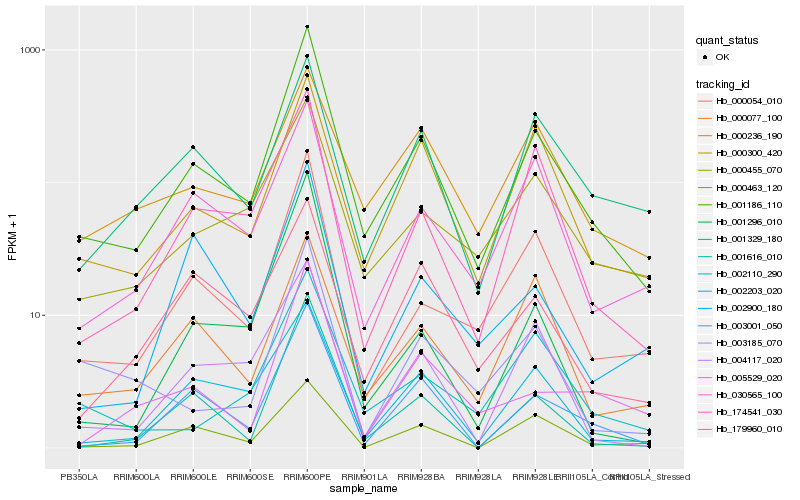
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001186_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 2 |
Hb_000054_010 |
0.1359715614 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003001_050 |
0.1466744993 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 4 |
Hb_000455_070 |
0.156193025 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 5 |
Hb_002203_020 |
0.1604606768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000300_420 |
0.1606446579 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_174541_030 |
0.1607404366 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 8 |
Hb_030565_100 |
0.1640399592 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 9 |
Hb_002110_290 |
0.1670331305 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_001616_010 |
0.1688153848 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 11 |
Hb_004117_020 |
0.1700085694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000077_100 |
0.1791140719 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005529_020 |
0.1792150243 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 14 |
Hb_001296_010 |
0.1793524759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001329_180 |
0.1812293537 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 16 |
Hb_000463_120 |
0.182749854 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 17 |
Hb_179960_010 |
0.1840539236 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 18 |
Hb_003185_070 |
0.1872954341 |
- |
- |
Plant natriuretic peptide A [Theobroma cacao] |
| 19 |
Hb_002900_180 |
0.1886496246 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 20 |
Hb_000236_190 |
0.1896486806 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |