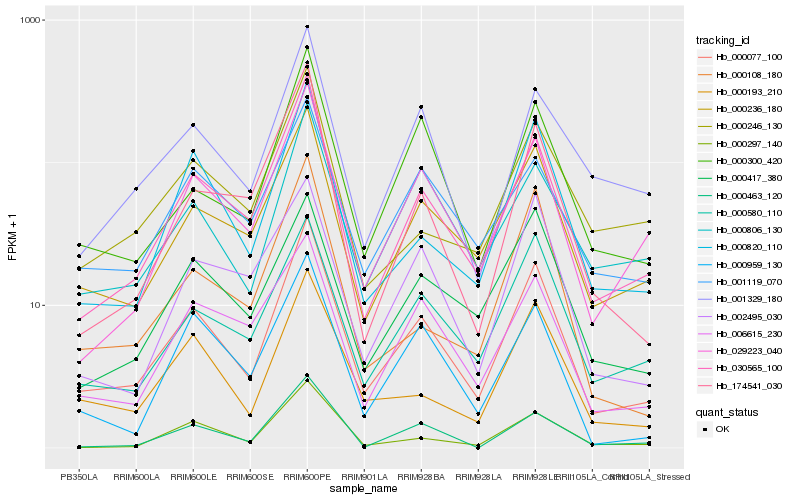
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_100 |
0.0 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_030565_100 |
0.0843304208 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000236_180 |
0.1079295788 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 4 |
Hb_029223_040 |
0.1190673615 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 5 |
Hb_000820_110 |
0.1197461654 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 6 |
Hb_000193_210 |
0.1219880835 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 7 |
Hb_000580_110 |
0.12340778 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 8 |
Hb_000300_420 |
0.1273879367 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000297_140 |
0.1339744478 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 10 |
Hb_001329_180 |
0.1386583421 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 11 |
Hb_000959_130 |
0.1389762801 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 12 |
Hb_000417_380 |
0.1396530365 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 13 |
Hb_006615_230 |
0.1404863713 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_174541_030 |
0.1409169161 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 15 |
Hb_000806_130 |
0.1415848025 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 16 |
Hb_000463_120 |
0.1437250702 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 17 |
Hb_001119_070 |
0.1451932736 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 18 |
Hb_000108_180 |
0.1452360701 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 19 |
Hb_002495_030 |
0.1483834365 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000246_130 |
0.153444265 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |